This is a preprint.
Differential escape of neutralizing antibodies by SARS-CoV-2 Omicron and pre-emergent sarbecoviruses
- PMID: 35233568
- PMCID: PMC8887082
- DOI: 10.21203/rs.3.rs-1362541/v1
Differential escape of neutralizing antibodies by SARS-CoV-2 Omicron and pre-emergent sarbecoviruses
Abstract
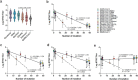
The SARS-CoV-2 B.1.1.529 lineage, Omicron variant, was first detected in November 2021 and carries 32 amino acid mutations in the spike protein (15 in RBD) and exhibits significant escape of neutralizing antibodies targeting the parental SARS-CoV-2 virus. Here, we performed a high-resolution multiplex (16-plex) surrogate virus neutralization assay covering all major SARS-CoV-2 variants and pre-emergent ACE2-binding sarbecoviruses against 20 different human serum panels from infected, vaccinated and hybrid immune individuals which had vaccine-breakthrough infections or infection followed by vaccination. Among all sarbecoviruses tested, we observed 1.1 to 4.7-, 2.3 to 10.3- and 0.7 to 33.3-fold reduction in neutralization activities to SARS-CoV-2 Beta, Omicron and SARS-CoV-1, respectively. Among the SARS-CoV-2 related sarbecoviruses, it is found that the genetically more distant bat RaTG13 and pangolin GX-P5L sarbecoviruses had less neutralization escape than Omicron. Our data suggest that the SARS-CoV-2 variants emerged from the changed immune landscape of human populations are more potent in escaping neutralizing antibodies, from infection or vaccination, than pre-emergent sarbecoviruses naturally evolved in animal populations with no or less immune selection pressure.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

