This is a preprint.
Type I interferon responsive microglia shape cortical development and behavior
- PMID: 35233577
- PMCID: PMC8887080
- DOI: 10.1101/2021.04.29.441889
Type I interferon responsive microglia shape cortical development and behavior
Update in
-
Type-I-interferon-responsive microglia shape cortical development and behavior.Cell. 2024 Apr 11;187(8):1936-1954.e24. doi: 10.1016/j.cell.2024.02.020. Epub 2024 Mar 14. Cell. 2024. PMID: 38490196 Free PMC article.
Abstract
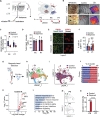
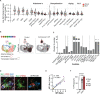
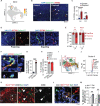
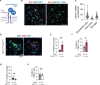
Microglia are brain resident phagocytes that can engulf synaptic components and extracellular matrix as well as whole neurons. However, whether there are unique molecular mechanisms that regulate these distinct phagocytic states is unknown. Here we define a molecularly distinct microglial subset whose function is to engulf neurons in the developing brain. We transcriptomically identified a cluster of Type I interferon (IFN-I) responsive microglia that expanded 20-fold in the postnatal day 5 somatosensory cortex after partial whisker deprivation, a stressor that accelerates neural circuit remodeling. In situ, IFN-I responsive microglia were highly phagocytic and actively engulfed whole neurons. Conditional deletion of IFN-I signaling (Ifnar1fl/fl) in microglia but not neurons resulted in dysmorphic microglia with stalled phagocytosis and an accumulation of neurons with double strand DNA breaks, a marker of cell stress. Conversely, exogenous IFN-I was sufficient to drive neuronal engulfment by microglia and restrict the accumulation of damaged neurons. IFN-I deficient mice had excess excitatory neurons in the developing somatosensory cortex as well as tactile hypersensitivity to whisker stimulation. These data define a molecular mechanism through which microglia engulf neurons during a critical window of brain development. More broadly, they reveal key homeostatic roles of a canonical antiviral signaling pathway in brain development.
Conflict of interest statement
Declaration of interests: The authors declare no competing interests.
Figures


References
-
- Arguello P. A. & Gogos J. A. Genetic and cognitive windows into circuit mechanisms of psychiatric disease. Trends Neurosci. 35, 3–13 (2012). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
