Outcome Prediction of Postanoxic Coma: A Comparison of Automated Electroencephalography Analysis Methods
- PMID: 35233717
- PMCID: PMC9343315
- DOI: 10.1007/s12028-022-01449-8
Outcome Prediction of Postanoxic Coma: A Comparison of Automated Electroencephalography Analysis Methods
Abstract
Background: To compare three computer-assisted quantitative electroencephalography (EEG) prediction models for the outcome prediction of comatose patients after cardiac arrest regarding predictive performance and robustness to artifacts.
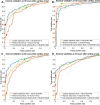
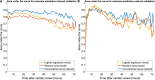
Methods: A total of 871 continuous EEGs recorded up to 3 days after cardiac arrest in intensive care units of five teaching hospitals in the Netherlands were retrospectively analyzed. Outcome at 6 months was dichotomized as "good" (Cerebral Performance Category 1-2) or "poor" (Cerebral Performance Category 3-5). Three prediction models were implemented: a logistic regression model using two quantitative features, a random forest model with nine features, and a deep learning model based on a convolutional neural network. Data from two centers were used for training and fivefold cross-validation (n = 663), and data from three other centers were used for external validation (n = 208). Model output was the probability of good outcome. Predictive performances were evaluated by using receiver operating characteristic analysis and the calculation of predictive values. Robustness to artifacts was evaluated by using an artifact rejection algorithm, manually added noise, and randomly flattened channels in the EEG.
Results: The deep learning network showed the best overall predictive performance. On the external test set, poor outcome could be predicted by the deep learning network at 24 h with a sensitivity of 54% (95% confidence interval [CI] 44-64%) at a false positive rate (FPR) of 0% (95% CI 0-2%), significantly higher than the logistic regression (sensitivity 33%, FPR 0%) and random forest models (sensitivity 13%, FPR, 0%) (p < 0.05). Good outcome at 12 h could be predicted by the deep learning network with a sensitivity of 78% (95% CI 52-100%) at a FPR of 12% (95% CI 0-24%) and by the logistic regression model with a sensitivity of 83% (95% CI 83-83%) at a FPR of 3% (95% CI 3-3%), both significantly higher than the random forest model (sensitivity 1%, FPR 0%) (p < 0.05). The results of the deep learning network were the least affected by the presence of artifacts, added white noise, and flat EEG channels.
Conclusions: A deep learning model outperformed logistic regression and random forest models for reliable, robust, EEG-based outcome prediction of comatose patients after cardiac arrest.
Keywords: Brain hypoxia; Cardiac arrest; Deep neural networks; Electroencephalography; Machine learning; Prognosis.
© 2022. The Author(s).
Conflict of interest statement
Dr. van Putten is co-founder of Clinical Science Systems, a supplier of electroencephalogram systems for Medisch Spectrum Twente. Dr. Blans received funding from Philips (ultrasound seminar). The remaining authors have disclosed that they do not have any conflicts of interest.
Figures



References
-
- Cloostermans MC, Van Meulen FB, Eertman CJ, et al. Continuous electroencephalography monitoring for early prediction of neurological outcome in postanoxic patients after cardiac arrest: a prospective cohort study. Crit Care Med. 2012;40(10):2867–2875. doi: 10.1097/CCM.0b013e31825b94f0. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

