ΔNp63-Senataxin circuit controls keratinocyte differentiation by promoting the transcriptional termination of epidermal genes
- PMID: 35235452
- PMCID: PMC8915885
- DOI: 10.1073/pnas.2104718119
ΔNp63-Senataxin circuit controls keratinocyte differentiation by promoting the transcriptional termination of epidermal genes
Abstract
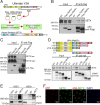
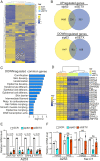
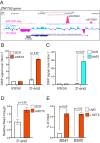
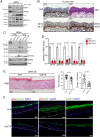
SignificanceΔNp63 is a master regulator of skin homeostasis since it finely controls keratinocyte differentiation and proliferation. Here, we provide cellular and molecular evidence demonstrating the functional role of a ΔNp63 interactor, the R-loop-resolving enzyme Senataxin (SETX), in fine-tuning keratinocyte differentiation. We found that SETX physically binds the p63 DNA-binding motif present in two early epidermal differentiation genes, Keratin 1 (KRT1) and ZNF750, facilitating R-loop removal over their 3' ends and thus allowing efficient transcriptional termination and gene expression. These molecular events translate into the inability of SETX-depleted keratinocytes to undergo the correct epidermal differentiation program. Remarkably, SETX is dysregulated in cutaneous squamous cell carcinoma, suggesting its potential involvement in the pathogenesis of skin disorders.
Keywords: Senataxin; p63; skin differentiation.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Candi E., Schmidt R., Melino G., The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6, 328–340 (2005). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

