To the proteome and beyond: advances in single-cell omics profiling for plant systems
- PMID: 35235661
- PMCID: PMC8825333
- DOI: 10.1093/plphys/kiab429
To the proteome and beyond: advances in single-cell omics profiling for plant systems
Abstract
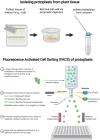
Recent advances in single-cell proteomics for animal systems could be adapted for plants to increase our understanding of plant development, response to stimuli, and cell-to-cell signaling.
Figures


References
-
- Beeck JOD, Pauwels J, Landuyt NV, Jacobs P, Malsche WD, Desmet G, Argentini A, Staes A, Martens L, Impens F, et al. (2019) A well-ordered nanoflow LC-MS/MS approach for proteome profiling using 200 cm long micro pillar array columns. bioRxiv 472134
-
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN (2005) Cell type–specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2: 615–619 - PubMed
-
- Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, et al. (2013) Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 10: 1093–1095 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

