The TΑp63/BCL2 axis represents a novel mechanism of clinical aggressiveness in chronic lymphocytic leukemia
- PMID: 35235952
- PMCID: PMC9043946
- DOI: 10.1182/bloodadvances.2021006348
The TΑp63/BCL2 axis represents a novel mechanism of clinical aggressiveness in chronic lymphocytic leukemia
Abstract
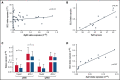
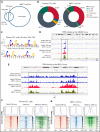
The TA-isoform of the p63 transcription factor (TAp63) has been reported to contribute to clinical aggressiveness in chronic lymphocytic leukemia (CLL) in a hitherto elusive way. Here, we sought to further understand and define the role of TAp63 in the pathophysiology of CLL. First, we found that elevated TAp63 expression levels are linked with adverse clinical outcomes, including disease relapse and shorter time-to-first treatment and overall survival. Next, prompted by the fact that TAp63 participates in an NF-κB/TAp63/BCL2 antiapoptotic axis in activated mature, normal B cells, we explored molecular links between TAp63 and BCL2 also in CLL. We documented a strong correlation at both the protein and the messenger RNA (mRNA) levels, alluding to the potential prosurvival role of TAp63. This claim was supported by inducible downregulation of TAp63 expression in the MEC1 CLL cell line using clustered regularly interspaced short palindromic repeats (CRISPR) system, which resulted in downregulation of BCL2 expression. Next, using chromatin immunoprecipitation (ChIP) sequencing, we examined whether BCL2 might constitute a transcriptional target of TAp63 and identified a significant binding profile of TAp63 in the BCL2 gene locus, across a genomic region previously characterized as a super enhancer in CLL. Moreover, we identified high-confidence TAp63 binding regions in genes mainly implicated in immune response and DNA-damage procedures. Finally, we found that upregulated TAp63 expression levels render CLL cells less responsive to apoptosis induction with the BCL2 inhibitor venetoclax. On these grounds, TAp63 appears to act as a positive modulator of BCL2, hence contributing to the antiapoptotic phenotype that underlies clinical aggressiveness and treatment resistance in CLL.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures



References
-
- Gonfloni S, Caputo V, Iannizzotto V. P63 in health and cancer. Int J Dev Biol. 2015;59(1-3):87-93. - PubMed
-
- Alexandrova EM, Moll UM. Role of p53 family members p73 and p63 in human hematological malignancies. Leuk Lymphoma. 2012;53(11):2116-2129. - PubMed
-
- Papakonstantinou N, Ntoufa S, Tsagiopoulou M, et al. . Integrated epigenomic and transcriptomic analysis reveals TP63 as a novel player in clinically aggressive chronic lymphocytic leukemia. Int J Cancer. 2019;144(11):2695-2706. - PubMed
-
- Lantner F, Starlets D, Gore Y, et al. . CD74 induces TAp63 expression leading to B-cell survival. Blood. 2007;110(13):4303-4311. - PubMed

