Human serum from SARS-CoV-2-vaccinated and COVID-19 patients shows reduced binding to the RBD of SARS-CoV-2 Omicron variant
- PMID: 35236358
- PMCID: PMC8890955
- DOI: 10.1186/s12916-022-02312-5
Human serum from SARS-CoV-2-vaccinated and COVID-19 patients shows reduced binding to the RBD of SARS-CoV-2 Omicron variant
Abstract
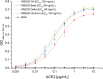
Background: The COVID-19 pandemic is caused by the betacoronavirus SARS-CoV-2. In November 2021, the Omicron variant was discovered and immediately classified as a variant of concern (VOC), since it shows substantially more mutations in the spike protein than any previous variant, especially in the receptor-binding domain (RBD). We analyzed the binding of the Omicron RBD to the human angiotensin-converting enzyme-2 receptor (ACE2) and the ability of human sera from COVID-19 patients or vaccinees in comparison to Wuhan, Beta, or Delta RBD variants.
Methods: All RBDs were produced in insect cells. RBD binding to ACE2 was analyzed by ELISA and microscale thermophoresis (MST). Similarly, sera from 27 COVID-19 patients, 81 vaccinated individuals, and 34 booster recipients were titrated by ELISA on RBDs from the original Wuhan strain, Beta, Delta, and Omicron VOCs. In addition, the neutralization efficacy of authentic SARS-CoV-2 wild type (D614G), Delta, and Omicron by sera from 2× or 3× BNT162b2-vaccinated persons was analyzed.
Results: Surprisingly, the Omicron RBD showed a somewhat weaker binding to ACE2 compared to Beta and Delta, arguing that improved ACE2 binding is not a likely driver of Omicron evolution. Serum antibody titers were significantly lower against Omicron RBD compared to the original Wuhan strain. A 2.6× reduction in Omicron RBD binding was observed for serum of 2× BNT162b2-vaccinated persons. Neutralization of Omicron SARS-CoV-2 was completely diminished in our setup.
Conclusion: These results indicate an immune escape focused on neutralizing antibodies. Nevertheless, a boost vaccination increased the level of anti-RBD antibodies against Omicron, and neutralization of authentic Omicron SARS-CoV-2 was at least partially restored. This study adds evidence that current vaccination protocols may be less efficient against the Omicron variant.
Keywords: Antibody titer; Beta variant (B.1.351); COVID-19; Delta variant (B.1.617.2); Human angiotensin-converting enzyme-2 receptor (ACE2); Omicron variant (B.1.1.529); Receptor-binding domain (RBD); SARS-CoV-2; Vaccination; Virus neutralization.
© 2022. The Author(s).
Conflict of interest statement
MS1, FB1, SS, PAH, SD, and MH are inventors on a patent application on blocking antibodies against SARS-CoV-2. SD and MH are co-founders and shareholders of CORAT Therapeutics GmbH, a company founded for clinical and regulatory development of COR-101, an antibody for the treatment of hospitalized COVID-19 patients. SD and EVW are co-founders and shareholders of Abcalis GmbH, a company producing antibodies for diagnostics of SARS-CoV-2.
Figures


References
-
- Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1..... Accessed 8 Dec 2021.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

