Gramicidin S and melittin: potential anti-viral therapeutic peptides to treat SARS-CoV-2 infection
- PMID: 35236909
- PMCID: PMC8891299
- DOI: 10.1038/s41598-022-07341-x
Gramicidin S and melittin: potential anti-viral therapeutic peptides to treat SARS-CoV-2 infection
Abstract
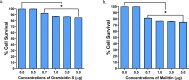
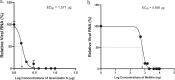
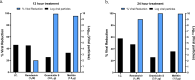
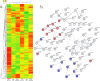
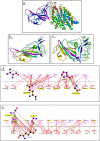
The COVID19 pandemic has led to multipronged approaches for treatment of the disease. Since de novo discovery of drugs is time consuming, repurposing of molecules is now considered as one of the alternative strategies to treat COVID19. Antibacterial peptides are being recognized as attractive candidates for repurposing to treat viral infections. In this study, we describe the anti-SARS-CoV-2 activity of the well-studied antibacterial peptides gramicidin S and melittin obtained from Bacillus brevis and bee venom respectively. The EC50 values for gramicidin S and melittin were 1.571 µg and 0.656 µg respectively based on in vitro antiviral assay. Significant decrease in the viral load as compared to the untreated group with no/very less cytotoxicity was observed. Both the peptides treated to the SARS-CoV-2 infected Vero cells showed viral clearance from 12 h onwards with a maximal viral clearance after 24 h post infection. Proteomics analysis indicated that more than 250 proteins were differentially regulated in the gramicidin S and melittin treated SARS-CoV-2 infected Vero cells against control SARS-CoV-2 infected Vero cells after 24 and 48 h post infection. The identified proteins were found to be associated in the metabolic and mRNA processing of the Vero cells post-treatment and infection. Both these peptides could be attractive candidates for repurposing to treat SARS-CoV-2 infection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 vaccines. JAMA. 2021;325:1318–1320. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

