Toll-Like Receptor Evolution: Does Temperature Matter?
- PMID: 35237266
- PMCID: PMC8882821
- DOI: 10.3389/fimmu.2022.812890
Toll-Like Receptor Evolution: Does Temperature Matter?
Abstract
Toll-like receptors (TLRs) recognize conserved pathogen-associated molecular patterns (PAMPs) and are an ancient and well-conserved group of pattern recognition receptors (PRRs). The isolation of the Antarctic continent and its unique teleost fish and microbiota prompted the present investigation into Tlr evolution. Gene homologues of tlr members in teleosts from temperate regions were present in the genome of Antarctic Nototheniidae and the non-Antarctic sister lineage Bovichtidae. Overall, in Nototheniidae apart from D. mawsoni, no major tlr gene family expansion or contraction occurred. Instead, lineage and species-specific changes in the ectodomain and LRR of Tlrs occurred, particularly in the Tlr11 superfamily that is well represented in fish. Positive selective pressure and associated sequence modifications in the TLR ectodomain and within the leucine-rich repeats (LRR), important for pathogen recognition, occurred in Tlr5, Tlr8, Tlr13, Tlr21, Tlr22, and Tlr23 presumably associated with the unique Antarctic microbiota. Exposure to lipopolysaccharide (Escherichia coli O111:B4) Gram negative bacteria did not modify tlr gene expression in N. rossii head-kidney or anterior intestine, although increased water temperature (+4°C) had a significant effect.
Keywords: Antarctic fish; TLR; cold temperature; evolution; immune challenge; innate immunity.
Copyright © 2022 Sousa, Fernandes, Cardoso, Wang, Zhai, Guerreiro, Chen, Canário and Power.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




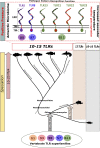
References
-
- Tort L, Balasch JC, Mackenzie S. Fish Immune System. A Crossroads Between Innate and Adaptive Responses. Inmunologia (2003) 22:277–86. doi: 10.1016/j.dci.2011.07.002 - DOI
-
- Uribe C, Folch H, Enriquez R, Moran G. Innate and Adaptive Immunity in Teleost Fish: A Review. Vet Med (2011) 56:486–503. doi: 10.17221/3294-VETMED - DOI
-
- Soulliere C, Dixon B. Immune System Organs of Bony Fishes. Ref Module Life Sci (2017) 1–5. doi: 10.1016/B978-0-12-809633-8.12179-X - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

