Circulating Long Non-Coding RNAs LINC00324 and LOC100507053 as Potential Liquid Biopsy Markers for Esophageal Squamous Cell Carcinoma: A Pilot Study
- PMID: 35237522
- PMCID: PMC8882835
- DOI: 10.3389/fonc.2022.823953
Circulating Long Non-Coding RNAs LINC00324 and LOC100507053 as Potential Liquid Biopsy Markers for Esophageal Squamous Cell Carcinoma: A Pilot Study
Abstract
Background: Despite the availability of advanced technology to detect and treat esophageal squamous cell carcinoma (ESCC), the 5-year survival rate of ESCC patients is still meager. Recently, long non-coding RNAs (lncRNAs) have emerged as essential players in the diagnosis and prognosis of various cancers.
Objective: This pilot study focused on identifying circulating lncRNAs as liquid biopsy markers for the ESCC.
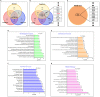
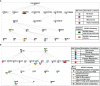
Methodology: We performed next-generation sequencing (NGS) to profile circulating lncRNAs in ESCC and healthy individuals' blood samples. The expression of the top five upregulated and top five downregulated lncRNAs were validated through quantitative real-time PCR (qRT-PCR), including samples used for the NGS. Later, we explored the diagnostic/prognostic potential of lncRNAs and their impact on the clinicopathological parameters of patients. To unravel the molecular target and pathways of identified lncRNAs, we utilized various bioinformatics tools such as lncRnome, RAID v2.0, Starbase, miRDB, TargetScan, Gene Ontology, and KEGG pathways.
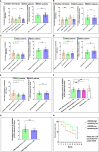
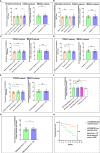
Results: Through NGS profiling, we obtained 159 upregulated, 137 downregulated, and 188 neutral lncRNAs in ESCC blood samples compared to healthy individuals. Among dysregulated lncRNAs, we observed LINC00324 significantly upregulated (2.11-fold; p-value = 0.0032) and LOC100507053 significantly downregulated (2.22-fold; p-value = 0.0001) in ESCC patients. Furthermore, we found LINC00324 and LOC100507053 could discriminate ESCC cancer patients' from non-cancer individuals with higher accuracy of Area Under the ROC Curve (AUC) = 0.627 and 0.668, respectively. The Kaplan-Meier and log-rank analysis revealed higher expression levels of LINC00324 and lower expression levels of LOC100507053 well correlated with the poor prognosis of ESCC patients with a Hazard ratio of LINC00324 = 2.48 (95% CI: 1.055 to 5.835) and Hazard ratio of LOC100507053 = 4.75 (95% CI: 2.098 to 10.76)]. Moreover, we also observed lncRNAs expression well correlated with the age (>50 years), gender (Female), alcohol, tobacco, and hot beverages consumers. Using bioinformatics tools, we saw miR-493-5p as the direct molecular target of LINC00324 and interacted with the MAPK signaling pathway in ESCC pathogenesis.
Conclusion: This pilot study suggests that circulating LINC00324 and LOC100507053 can be used as a liquid biopsy marker of ESCC; however, multicentric studies are still warranted.
Keywords: ESCC; LncRNA; biomarkers; esophageal squamous cell carcinoma; long non-coding RNA; next generation sequencing.
Copyright © 2022 Sharma, Barwal, Khandelwal, Rana, Rana, Singh and Jain.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

