Amino acid sensor conserved from bacteria to humans
- PMID: 35238638
- PMCID: PMC8915833
- DOI: 10.1073/pnas.2110415119
Amino acid sensor conserved from bacteria to humans
Abstract
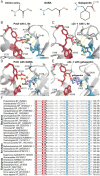
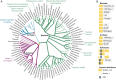
SignificanceAmino acids are the building blocks of life and important signaling molecules. Despite their common structure, no universal mechanism for amino acid recognition by cellular receptors is currently known. We discovered a simple motif, which binds amino acids in various receptor proteins from all major life-forms. In humans, this motif is found in subunits of calcium channels that are implicated in pain and neurodevelopmental disorders. Our findings suggest that γ-aminobutyric acid-derived drugs bind to the same motif in human proteins that binds natural ligands in bacterial receptors, thus enabling future improvement of important drugs.
Keywords: evolution; gabapentin; ion channels; serine/threonine kinases; signal transduction.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

