Deep learning guided optimization of human antibody against SARS-CoV-2 variants with broad neutralization
- PMID: 35238654
- PMCID: PMC8931377
- DOI: 10.1073/pnas.2122954119
Deep learning guided optimization of human antibody against SARS-CoV-2 variants with broad neutralization
Abstract
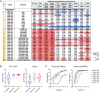
SignificanceSARS-CoV-2 continues to evolve through emerging variants, more frequently observed with higher transmissibility. Despite the wide application of vaccines and antibodies, the selection pressure on the Spike protein may lead to further evolution of variants that include mutations that can evade immune response. To catch up with the virus's evolution, we introduced a deep learning approach to redesign the complementarity-determining regions (CDRs) to target multiple virus variants and obtained an antibody that broadly neutralizes SARS-CoV-2 variants.
Keywords: SARS-CoV-2 variants; broadly neutralizing antibodies; computational biology; deep learning; geometric neural networks.
Conflict of interest statement
Competing interest statement: A patent has been filed for the optimized antibodies targeting SARS-CoV-2. J.P., J.M., and S.L. are employees of Helixon.
Figures

Similar articles
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. Elife. 2020. PMID: 33112236 Free PMC article.
-
Elicitation of Broadly Neutralizing Antibodies against B.1.1.7, B.1.351, and B.1.617.1 SARS-CoV-2 Variants by Three Prototype Strain-Derived Recombinant Protein Vaccines.Viruses. 2021 Jul 22;13(8):1421. doi: 10.3390/v13081421. Viruses. 2021. PMID: 34452287 Free PMC article.
-
Cross-Neutralization of Emerging SARS-CoV-2 Variants of Concern by Antibodies Targeting Distinct Epitopes on Spike.mBio. 2021 Dec 21;12(6):e0297521. doi: 10.1128/mBio.02975-21. Epub 2021 Nov 16. mBio. 2021. PMID: 34781736 Free PMC article. Clinical Trial.
-
Comprehensive Overview of Broadly Neutralizing Antibodies against SARS-CoV-2 Variants.Viruses. 2024 Jun 1;16(6):900. doi: 10.3390/v16060900. Viruses. 2024. PMID: 38932192 Free PMC article. Review.
-
Structural Immunology of SARS-CoV-2.Immunol Rev. 2025 Jan;329(1):e13431. doi: 10.1111/imr.13431. Epub 2024 Dec 27. Immunol Rev. 2025. PMID: 39731211 Free PMC article. Review.
Cited by
-
Machine Learning-Guided Protein Engineering.ACS Catal. 2023 Oct 13;13(21):13863-13895. doi: 10.1021/acscatal.3c02743. eCollection 2023 Nov 3. ACS Catal. 2023. PMID: 37942269 Free PMC article. Review.
-
De novo generation of SARS-CoV-2 antibody CDRH3 with a pre-trained generative large language model.Nat Commun. 2024 Aug 10;15(1):6867. doi: 10.1038/s41467-024-50903-y. Nat Commun. 2024. PMID: 39127753 Free PMC article.
-
Single-cell spatiotemporal analysis reveals alveolar dendritic cell-T cell immunity hubs defending against pulmonary infection.Cell Discov. 2024 Oct 16;10(1):103. doi: 10.1038/s41421-024-00733-5. Cell Discov. 2024. PMID: 39414763 Free PMC article.
-
Modeling the competitive transmission of the Omicron strain and Delta strain of COVID-19.J Math Anal Appl. 2023 Oct 15;526(2):127283. doi: 10.1016/j.jmaa.2023.127283. Epub 2023 Mar 31. J Math Anal Appl. 2023. PMID: 37035507 Free PMC article.
-
RLEAAI: improving antibody-antigen interaction prediction using protein language model and sequence order information.Brief Bioinform. 2025 May 1;26(3):bbaf238. doi: 10.1093/bib/bbaf238. Brief Bioinform. 2025. PMID: 40462512 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

