Mucus, commensals, and the immune system
- PMID: 35239459
- PMCID: PMC8903774
- DOI: 10.1080/19490976.2022.2041342
Mucus, commensals, and the immune system
Abstract
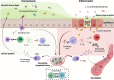
The immune system in the large intestine is separated from commensal microbes and comparatively rare enteric pathogens by a monolayer of diverse epithelial cells overlaid with a compact and adherent inner mucus layer and a looser outer mucus layer. Microorganisms, collectively referred to as the mucus-associated (MA) microbiota, physically inhabit this mucus barrier, resulting in a dynamic and incessant dialog to maintain both spatial segregation and immune tolerance. Recent major findings reveal novel features of the crosstalk between the immune system and mucus-associated bacteria in health and disease, as well as disease-related peripheral immune signatures indicative of host responses to these organisms. In this brief review, we integrate these novel observations into our overall understanding of host-microbiota mutualism at the colonic mucosal border and speculate on the significance of this emerging knowledge for our understanding of the prevention, development, and progression of chronic intestinal inflammation.
Keywords: Colon mucus layer; T-dependent; T-independent; anti-commensal IgA; anti-commensal IgG; flagellin; lachnospiraceae; mucus-associated bacteria.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS.. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect Immun. 2004;72:1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004. - DOI - PMC - PubMed
-
- Ravcheev DA, Thiele I. Comparative genomic analysis of the human gut microbiome reveals a broad distribution of metabolic pathways for the degradation of host-synthetized mucin glycans and utilization of mucin-derived monosaccharides. Front Genet. 2017;8:111. doi: 10.3389/fgene.2017.00111. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
