ER-PM membrane contact site regulation by yeast ORPs and membrane stress pathways
- PMID: 35239652
- PMCID: PMC8923467
- DOI: 10.1371/journal.pgen.1010106
ER-PM membrane contact site regulation by yeast ORPs and membrane stress pathways
Abstract
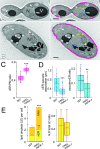
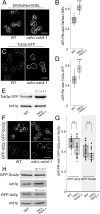
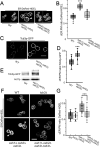
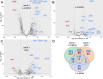
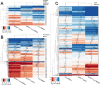
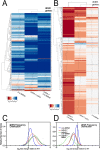
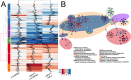
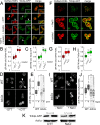
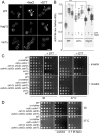
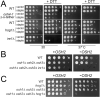
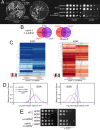
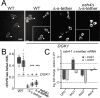
In yeast, at least seven proteins (Ice2p, Ist2p, Scs2/22p, Tcb1-Tcb3p) affect cortical endoplasmic reticulum (ER) tethering and contact with the plasma membrane (PM). In Δ-super-tether (Δ-s-tether) cells that lack these tethers, cortical ER-PM association is all but gone. Yeast OSBP homologue (Osh) proteins are also implicated in membrane contact site (MCS) assembly, perhaps as subunits for multicomponent tethers, though their function at MCSs involves intermembrane lipid transfer. Paradoxically, when analyzed by fluorescence and electron microscopy, the elimination of the OSH gene family does not reduce cortical ER-PM association but dramatically increases it. In response to the inactivation of all Osh proteins, the yeast E-Syt (extended-synaptotagmin) homologue Tcb3p is post-transcriptionally upregulated thereby generating additional Tcb3p-dependent ER-PM MCSs for recruiting more cortical ER to the PM. Although the elimination of OSH genes and the deletion of ER-PM tether genes have divergent effects on cortical ER-PM association, both elicit the Environmental Stress Response (ESR). Through comparisons of transcriptomic profiles of cells lacking OSH genes or ER-PM tethers, changes in ESR expression are partially manifested through the induction of the HOG (high-osmolarity glycerol) PM stress pathway or the ER-specific UPR (unfolded protein response) pathway, respectively. Defects in either UPR or HOG pathways also increase ER-PM MCSs, and expression of extra "artificial ER-PM membrane staples" rescues growth of UPR mutants challenged with lethal ER stress. Transcriptome analysis of OSH and Δ-s-tether mutants also revealed dysregulation of inositol-dependent phospholipid gene expression, and the combined lethality of osh4Δ and Δ-s-tether mutations is suppressed by overexpression of the phosphatidic acid biosynthetic gene, DGK1. These findings establish that the Tcb3p tether is induced by ER and PM stresses and ER-PM MCSs augment responses to membrane stresses, which are integrated through the broader ESR pathway.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

