SARS-CoV-2 Nucleocapsid Protein Targets a Conserved Surface Groove of the NTF2-like Domain of G3BP1
- PMID: 35240128
- PMCID: PMC8882607
- DOI: 10.1016/j.jmb.2022.167516
SARS-CoV-2 Nucleocapsid Protein Targets a Conserved Surface Groove of the NTF2-like Domain of G3BP1
Abstract
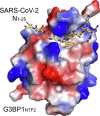
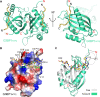
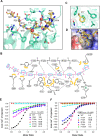
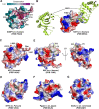
Stress granule (SG) formation mediated by Ras GTPase-activating protein-binding protein 1 (G3BP1) constitutes a key obstacle for viral replication, which makes G3BP1 a frequent target for viruses. For instance, the SARS-CoV-2 nucleocapsid (N) protein interacts with G3BP1 directly to suppress SG assembly and promote viral production. However, the molecular basis for the SARS-CoV-2 N - G3BP1 interaction remains elusive. Here we report biochemical and structural analyses of the SARS-CoV-2 N - G3BP1 interaction, revealing differential contributions of various regions of SARS-CoV-2 N to G3BP1 binding. The crystal structure of the NTF2-like domain of G3BP1 (G3BP1NTF2) in complex with a peptide derived from SARS-CoV-2 N (residues 1-25, N1-25) reveals that SARS-CoV-2 N1-25 occupies a conserved surface groove of G3BP1NTF2 via surface complementarity. We show that a φ-x-F (φ, hydrophobic residue) motif constitutes the primary determinant for G3BP1NTF2-targeting proteins, while the flanking sequence underpins diverse secondary interactions. We demonstrate that mutation of key interaction residues of the SARS-CoV-2 N1-25 - G3BP1NTF2 complex leads to disruption of the SARS-CoV-2 N - G3BP1 interaction in vitro. Together, these results provide a molecular basis of the strain-specific interaction between SARS-CoV-2 N and G3BP1, which has important implications for the development of novel therapeutic strategies against SARS-CoV-2 infection.
Keywords: NTF2-like domain; SARS-CoV-2 nucleocapsid protein; pathogen-host interaction; ras GTPase-activating protein-binding protein 1; stress granule.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

