Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy
- PMID: 35241090
- PMCID: PMC8892709
- DOI: 10.1186/s12943-021-01487-4
Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy
Abstract
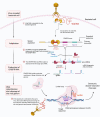
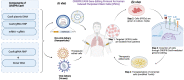
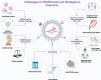
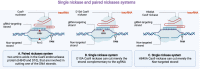
CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats-associated protein 9) shows the opportunity to treat a diverse array of untreated various genetic and complicated disorders. Therapeutic genome editing processes that target disease-causing genes or mutant genes have been greatly accelerated in recent years as a consequence of improvements in sequence-specific nuclease technology. However, the therapeutic promise of genome editing has yet to be explored entirely, many challenges persist that increase the risk of further mutations. Here, we highlighted the main challenges facing CRISPR/Cas9-based treatments and proposed strategies to overcome these limitations, for further enhancing this revolutionary novel therapeutics to improve long-term treatment outcome human health.
Keywords: CRISPR; Cancer therapy; Cas9; Gene editing; Gene modification challenges.
© 2022. The Author(s).
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures




References
-
- Lortet-Tieulent J, Georges D, Bray F, Vaccarella S. Profiling global cancer incidence and mortality by socioeconomic development. Int J Cancer. 2020;147(11):3029–36. - PubMed
-
- Arunachalam SS, Shetty AP, Panniyadi N, Meena C, Kumari J, Rani B, et al. Study on knowledge of chemotherapy’s adverse effects and their self-care ability to manage - The cancer survivors impact. Clin Epidemiol Glob Health. 2021;11:100765.
-
- Sachdeva M, Sachdeva N, Pal M, Gupta N, Khan IA, Majumdar M, et al. CRISPR/Cas9: molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Ther. 2015;22(11):509–17. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

