An orally available, brain penetrant, small molecule lowers huntingtin levels by enhancing pseudoexon inclusion
- PMID: 35241644
- PMCID: PMC8894458
- DOI: 10.1038/s41467-022-28653-6
An orally available, brain penetrant, small molecule lowers huntingtin levels by enhancing pseudoexon inclusion
Abstract
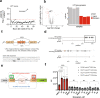
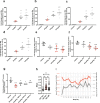
Huntington's Disease (HD) is a progressive neurodegenerative disorder caused by CAG trinucleotide repeat expansions in exon 1 of the huntingtin (HTT) gene. The mutant HTT (mHTT) protein causes neuronal dysfunction, causing progressive motor, cognitive and behavioral abnormalities. Current treatments for HD only alleviate symptoms, but cerebral spinal fluid (CSF) or central nervous system (CNS) delivery of antisense oligonucleotides (ASOs) or virus vectors expressing RNA-induced silencing (RNAi) moieties designed to induce mHTT mRNA lowering have progressed to clinical trials. Here, we present an alternative disease modifying therapy the orally available, brain penetrant small molecule branaplam. By promoting inclusion of a pseudoexon in the primary transcript, branaplam lowers mHTT protein levels in HD patient cells, in an HD mouse model and in blood samples from Spinal Muscular Atrophy (SMA) Type I patients dosed orally for SMA (NCT02268552). Our work paves the way for evaluating branaplam's utility as an HD therapy, leveraging small molecule splicing modulators to reduce expression of dominant disease genes by driving pseudoexon inclusion.
© 2022. The Author(s).
Conflict of interest statement
A.R., B.B., C.G.K., D.T., G.R., J.H.C. J.K., M.B., M.H., N.D., N.R., R.D., R.S., T.F., T.P., and Y.S. are (or were at the time the research was conducted) employees of Novartis. B.L.D. and A.M.M. have sponsored research from Novartis.
Figures


References
-
- MacDonald, M.E. et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell72, 971–983 (1993). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

