Cytoskeletal dynamics regulates stromal invasion behavior of distinct liver cancer subtypes
- PMID: 35241781
- PMCID: PMC8894393
- DOI: 10.1038/s42003-022-03121-5
Cytoskeletal dynamics regulates stromal invasion behavior of distinct liver cancer subtypes
Abstract
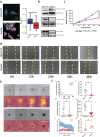
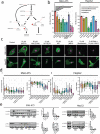
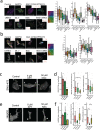
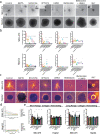
Drug treatment against liver cancer has limited efficacy due to heterogeneous response among liver cancer subtypes. In addition, the functional biophysical phenotypes which arise from this heterogeneity and contribute to aggressive invasive behavior remain poorly understood. This study interrogated how heterogeneity in liver cancer subtypes contributes to differences in invasive phenotypes and drug response. Utilizing histological analysis, quantitative 2D invasion metrics, reconstituted 3D hydrogels, and bioinformatics, our study linked cytoskeletal dynamics to differential invasion profiles and drug resistance in liver cancer subtypes. We investigated cytoskeletal regulation in 2D and 3D culture environments using two liver cancer cell lines, SNU-475 and HepG2, chosen for their distinct cytoskeletal features and invasion profiles. For SNU-475 cells, a model for aggressive liver cancer, many cytoskeletal inhibitors abrogated 2D migration but only some suppressed 3D migration. For HepG2 cells, cytoskeletal inhibition did not significantly affect 3D migration but did affect proliferative capabilities and spheroid core growth. This study highlights cytoskeleton driven phenotypic variation, their consequences and coexistence within the same tumor, as well as efficacy of targeting biophysical phenotypes that may be masked in traditional screens against tumor growth.
© 2022. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: B.E.E. is a cofounder of Osmol Therapeutics, a company that is targeting NCS1 for therapeutic purposes. The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

