Cysteine dependence of Lactobacillus iners is a potential therapeutic target for vaginal microbiota modulation
- PMID: 35241796
- PMCID: PMC10473153
- DOI: 10.1038/s41564-022-01070-7
Cysteine dependence of Lactobacillus iners is a potential therapeutic target for vaginal microbiota modulation
Abstract
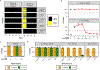
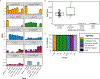
Vaginal microbiota composition affects many facets of reproductive health. Lactobacillus iners-dominated microbial communities are associated with poorer outcomes, including higher risk of bacterial vaginosis (BV), compared with vaginal microbiota rich in L. crispatus. Unfortunately, standard-of-care metronidazole therapy for BV typically results in dominance of L. iners, probably contributing to post-treatment relapse. Here we generate an L. iners isolate collection comprising 34 previously unreported isolates from 14 South African women with and without BV and 4 previously unreported isolates from 3 US women. We also report an associated genome catalogue comprising 1,218 vaginal Lactobacillus isolate genomes and metagenome-assembled genomes from >300 women across 4 continents. We show that, unlike L. crispatus, L. iners growth is dependent on L-cysteine in vitro and we trace this phenotype to the absence of canonical cysteine biosynthesis pathways and a restricted repertoire of cysteine-related transport mechanisms. We further show that cysteine concentrations in cervicovaginal lavage samples correlate with Lactobacillus abundance in vivo and that cystine uptake inhibitors selectively inhibit L. iners growth in vitro. Combining an inhibitor with metronidazole promotes L. crispatus dominance of defined BV-like communities in vitro by suppressing L. iners growth. Our findings enable a better understanding of L. iners biology and suggest candidate treatments to modulate the vaginal microbiota to improve reproductive health for women globally.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Author Contributions
S.M.B. and D.S.K. conceived the overall study and guided it throughout with input from B.M.W., E.P.B., and C.M.M.; S.M.B., N.A.M., and J.K.R. performed primary bacterial isolations; S.M.B., N.A.M., J.F.F., B.M.W., A.J.M., X.W., N.C., and C.M.M. contributed to media design and production and/or bacterial growth and inhibition experiments; B.M.W. and E.P.B. synthesized labeled glutathione; B.M.W., S.M.B., N.A.M., and E.P.B. designed, performed, and/or analyzed measurements of media composition and isotopic tracing experiments; S.M.B., N.A.M., and J.X. performed nucleic acid extractions and sequencing; S.M.B. performed bacterial 16S rRNA gene sequencing analysis; M.R.H. and D.A.R. performed bacterial isolate genomic and metagenomic sequence analysis and assembly, genome catalog development, and phylogenetic reconstructions; S.M.B., M.R.H., F.A.H., and B.M.W. conceived and/or performed genomic pathway analysis; S.M.B. and A.B.A. performed analysis of
Competing interests
All authors declare no competing interests.
Figures

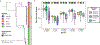


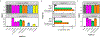





Comment in
-
Reconceptualizing bacterial vaginosis treatment.Nat Microbiol. 2022 Mar;7(3):365-366. doi: 10.1038/s41564-022-01063-6. Nat Microbiol. 2022. PMID: 35241797 No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

