Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease
- PMID: 35241825
- PMCID: PMC9005345
- DOI: 10.1038/s41588-021-01006-7
Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease
Abstract
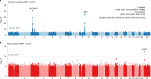
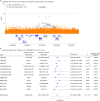
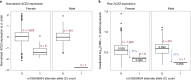
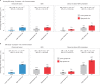
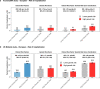
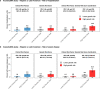
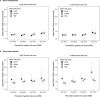
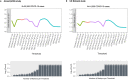
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) enters human host cells via angiotensin-converting enzyme 2 (ACE2) and causes coronavirus disease 2019 (COVID-19). Here, through a genome-wide association study, we identify a variant (rs190509934, minor allele frequency 0.2-2%) that downregulates ACE2 expression by 37% (P = 2.7 × 10-8) and reduces the risk of SARS-CoV-2 infection by 40% (odds ratio = 0.60, P = 4.5 × 10-13), providing human genetic evidence that ACE2 expression levels influence COVID-19 risk. We also replicate the associations of six previously reported risk variants, of which four were further associated with worse outcomes in individuals infected with the virus (in/near LZTFL1, MHC, DPP9 and IFNAR2). Lastly, we show that common variants define a risk score that is strongly associated with severe disease among cases and modestly improves the prediction of disease severity relative to demographic and clinical factors alone.
© 2022. The Author(s).
Conflict of interest statement
J.E.H., J.A.K., A.D., D. Sharma, N.B, A.Y., A.M., R.L., E.M., X.B., D. Sun, F.S.P.K., J.D.B., C.O.D., A.J.M., D.A.T., A.H.L., J. Mbatchou, K.W., L.G., S.E.M, H.M.K., L.D., E.S., M.J., S.B., K.S, W.J.S., A.R.S., A.E.L., J. Marchini, J.D.O., L.H., M.N.C., J.G.R., A. Baras, G.R.A. and M.A.R.F. are current and/or former employees and/or stockholders of RGC or Regeneron Pharmaceuticals. G.H.L.R., M.V.C., D.S.P., S.C.K. A. Baltzell, A.R.G., S.R.M., R.P., M.Z., K.A.R., E.L.H. and C.A.B. are current and/or former employees of AncestryDNA and may hold equity in AncestryDNA. The other authors declare no competing interests.
Figures



Update of
-
Genome-wide analysis in 756,646 individuals provides first genetic evidence that ACE2 expression influences COVID-19 risk and yields genetic risk scores predictive of severe disease.medRxiv [Preprint]. 2021 Jun 10:2020.12.14.20248176. doi: 10.1101/2020.12.14.20248176. medRxiv. 2021. Update in: Nat Genet. 2022 Apr;54(4):382-392. doi: 10.1038/s41588-021-01006-7. PMID: 33619501 Free PMC article. Updated. Preprint.
Comment in
-
Understanding COVID-19 through genome-wide association studies.Nat Genet. 2022 Apr;54(4):368-369. doi: 10.1038/s41588-021-00985-x. Nat Genet. 2022. PMID: 35410380 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

