Stem cell architecture drives myelodysplastic syndrome progression and predicts response to venetoclax-based therapy
- PMID: 35241842
- PMCID: PMC8938266
- DOI: 10.1038/s41591-022-01696-4
Stem cell architecture drives myelodysplastic syndrome progression and predicts response to venetoclax-based therapy
Erratum in
-
Author Correction: Stem cell architecture drives myelodysplastic syndrome progression and predicts response to venetoclax-based therapy.Nat Med. 2022 May;28(5):1097. doi: 10.1038/s41591-022-01827-x. Nat Med. 2022. PMID: 35484266 Free PMC article. No abstract available.
Abstract
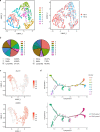
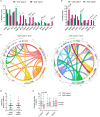
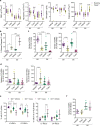
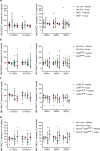
Myelodysplastic syndromes (MDS) are heterogeneous neoplastic disorders of hematopoietic stem cells (HSCs). The current standard of care for patients with MDS is hypomethylating agent (HMA)-based therapy; however, almost 50% of MDS patients fail HMA therapy and progress to acute myeloid leukemia, facing a dismal prognosis due to lack of approved second-line treatment options. As cancer stem cells are the seeds of disease progression, we investigated the biological properties of the MDS HSCs that drive disease evolution, seeking to uncover vulnerabilities that could be therapeutically exploited. Through integrative molecular profiling of HSCs and progenitor cells in large patient cohorts, we found that MDS HSCs in two distinct differentiation states are maintained throughout the clinical course of the disease, and expand at progression, depending on recurrent activation of the anti-apoptotic regulator BCL-2 or nuclear factor-kappa B-mediated survival pathways. Pharmacologically inhibiting these pathways depleted MDS HSCs and reduced tumor burden in experimental systems. Further, patients with MDS who progressed after failure to frontline HMA therapy and whose HSCs upregulated BCL-2 achieved improved clinical responses to venetoclax-based therapy in the clinical setting. Overall, our study uncovers that HSC architectures in MDS are potential predictive biomarkers to guide second-line treatments after HMA failure. These findings warrant further investigation of HSC-specific survival pathways to identify new therapeutic targets of clinical potential in MDS.
© 2022. The Author(s).
Conflict of interest statement
S.C. and M.A. report research funding from AbbVie and Amgen. J.P.I. and J.D.L. worked at AbbVie at the time of the study; G.G.-M. reports clinical funding from AbbVie and Amgen. All other authors report no competing interests related to this study.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

