Pan-Cancer Analysis Predicts FOXS1 as a Key Target in Prognosis and Tumor Immunotherapy
- PMID: 35241932
- PMCID: PMC8887970
- DOI: 10.2147/IJGM.S354195
Pan-Cancer Analysis Predicts FOXS1 as a Key Target in Prognosis and Tumor Immunotherapy
Abstract
Purpose: Only a few studies have reported the role of FOXS1, a transcriptional factor, in the tumor development process. In this article, we investigate the function of FOXS1 in distinct neoplastic development and the tumor immune microenvironment (TIME).
Patients and methods: The latent roles of FOXS1 in various tumors were prospected based on TCGA, GTEx, CCLE, GEPIA2, cBioPortal, TIMER, ImmuCellAI databases, GSVA datasets, GSEA datasets, and R packages. The expression difference, gene alteration, clinical characteristics, prognostic values, biological mechanism, potential pathways, tumor microenvironment, and immune cell infiltration related to FOXS1 were appraised.
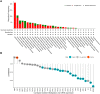
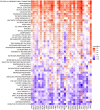
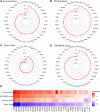
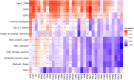
Results: FOXS1 was strongly expressed in pan-cancer, and this gene was associated with low survival rates. FOXS1 was linked to many pathways that are cancer-promoting and immune-related. The expression of this transcriptional factor in cancers was positively related to immune cell infiltration, especially M2-like macrophages and Treg cells. In addition to that, FOXS1 demonstrated a positive relationship with many immune-suppression genes, such as TGFB1 and ARORA2A.
Conclusion: Our study identified an oncogenic effect of FOXS1, which may play a vital role as a prognosticative biological marker in pan-cancer. Exorbitant expression of FOXS1 is associated with high TAMs and Treg cells infiltration. These cells have an immunosuppressive function and promote the development of the immunosuppressive tumor microenvironment. The research of FOXS1 provided a potential drug target for tumor immunotherapy.
Keywords: FOXS1; M2-like macrophages; Treg cells; biological marker; immunosuppressive microenvironment; pan-cancer.
© 2022 Liu et al.
Conflict of interest statement
The authors do not have any conflicts of interest to report for this work.
Figures





References
-
- Shrihari TG. Innate and adaptive immune cells in tumor microenvironment. Gulf J Oncolog. 2021;1(35):77–81. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

