Comparative Analysis of Intestinal Microflora Between Two Developmental Stages of Rimicaris kairei, a Hydrothermal Shrimp From the Central Indian Ridge
- PMID: 35242112
- PMCID: PMC8886129
- DOI: 10.3389/fmicb.2021.802888
Comparative Analysis of Intestinal Microflora Between Two Developmental Stages of Rimicaris kairei, a Hydrothermal Shrimp From the Central Indian Ridge
Abstract
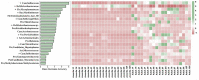
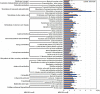
Despite extreme physical and chemical characteristics, deep-sea hydrothermal vents provide a place for fauna survival and reproduction. The symbiotic relationship of chemotrophic microorganisms has been investigated in the gill of Rimicaris exoculata, which are endemic to the hydrothermal vents of the Mid-Atlantic Ridge. However, only a few studies have examined intestinal symbiosis. Here, we studied the intestinal fauna in juvenile and adult Rimicaris kairei, another species in the Rimicaris genus that was originally discovered at the Kairei and Edmond hydrothermal vent fields in the Central Indian Ridge. The results showed that there were significant differences between juvenile and adult gut microbiota in terms of species richness, diversity, and evenness. The values of Chao1, observed species, and ASV rarefaction curves indicated almost four times the number of species in adults compared to juveniles. In juveniles, the most abundant phylum was Deferribacterota, at 80%, while in adults, Campilobacterota was the most abundant, at 49%. Beta diversity showed that the intestinal communities of juveniles and adults were clearly classified into two clusters based on the evaluations of Bray-Curtis and weighted UniFrac distance matrices. Deferribacteraceae and Sulfurovum were the main featured bacteria contributing to the difference. Moreover, functional prediction for all of the intestinal microbiota showed that the pathways related to ansamycin synthesis, branched-chain amino acid biosynthesis, lipid metabolism, and cell motility appeared highly abundant in juveniles. However, for adults, the most abundant pathways were those of sulfur transfer, carbohydrate, and biotin metabolism. Taken together, these results indicated large differences in intestinal microbial composition and potential functions between juvenile and adult vent shrimp (R. kairei), which may be related to their physiological needs at different stages of development.
Keywords: Deferribacteraceae; Rimicaris kairei; Sulfurovum; hydrothermal vent; intestine microflora.
Copyright © 2022 Qi, Lian, Zhu, Shi and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Symbiont Community Composition in Rimicaris kairei Shrimps from Indian Ocean Vents with Notes on Mineralogy.Appl Environ Microbiol. 2022 Apr 26;88(8):e0018522. doi: 10.1128/aem.00185-22. Epub 2022 Apr 11. Appl Environ Microbiol. 2022. PMID: 35404070 Free PMC article.
-
Genomic evidence for the first symbiotic Deferribacterota, a novel gut symbiont from the deep-sea hydrothermal vent shrimp Rimicaris kairei.Front Microbiol. 2023 Jun 29;14:1179935. doi: 10.3389/fmicb.2023.1179935. eCollection 2023. Front Microbiol. 2023. PMID: 37455748 Free PMC article.
-
"Candidatus Desulfobulbus rimicarensis," an Uncultivated Deltaproteobacterial Epibiont from the Deep-Sea Hydrothermal Vent Shrimp Rimicaris exoculata.Appl Environ Microbiol. 2020 Apr 1;86(8):e02549-19. doi: 10.1128/AEM.02549-19. Print 2020 Apr 1. Appl Environ Microbiol. 2020. PMID: 32060020 Free PMC article.
-
A new species of the genus Rimicaris (Alvinocarididae: Caridea: Decapoda) from the active hydrothermal vent field, "Kairei Field," on the Central Indian Ridge, the Indian Ocean.Zoolog Sci. 2002 Oct;19(10):1167-74. doi: 10.2108/zsj.19.1167. Zoolog Sci. 2002. PMID: 12426479
-
De novo transcriptome assembly of the deep-sea hydrothermal vent, shrimp Rimicaris exoculate (Crustacea: Decapoda), from the south Mid-Atlantic Ridge.Mar Genomics. 2021 Dec;60:100876. doi: 10.1016/j.margen.2021.100876. Epub 2021 May 3. Mar Genomics. 2021. PMID: 33958310
Cited by
-
Comparative analysis of esophageal gland microbes between two body sizes of Gigantopelta aegis, a hydrothermal snail from the Southwest Indian Ridge.Microbiol Spectr. 2025 Apr;13(4):e0295924. doi: 10.1128/spectrum.02959-24. Epub 2025 Feb 24. Microbiol Spectr. 2025. PMID: 39992146 Free PMC article.
-
The Application and Limitations of Promising Biological Therapies in Livestock Production under the Context of Antibiotic Restrictions.Probiotics Antimicrob Proteins. 2025 Aug 14. doi: 10.1007/s12602-025-10705-0. Online ahead of print. Probiotics Antimicrob Proteins. 2025. PMID: 40810777 Review.
-
Metabolomic and metagenomic analyses of the Chinese mitten crab Eriocheir sinensis after challenge with Metschnikowia bicuspidata.Front Microbiol. 2022 Sep 23;13:990737. doi: 10.3389/fmicb.2022.990737. eCollection 2022. Front Microbiol. 2022. PMID: 36212869 Free PMC article.
-
Symbioses of alvinocaridid shrimps from the South West Pacific: No chemosymbiotic diets but conserved gut microbiomes.Environ Microbiol Rep. 2023 Dec;15(6):614-630. doi: 10.1111/1758-2229.13201. Epub 2023 Sep 26. Environ Microbiol Rep. 2023. PMID: 37752716 Free PMC article.
-
Spatial variation of skin-associated microbiota in a green salamander metapopulation.Sci Rep. 2025 Jul 9;15(1):24738. doi: 10.1038/s41598-025-05305-5. Sci Rep. 2025. PMID: 40634333 Free PMC article.
References
-
- Apremont V., Cambon-Bonavita M. A., Cueff-Gauchard V., François D., Pradillon F., Corbari L., et al. (2018). Gill chamber and gut microbial communities of the hydrothermal shrimp Rimicaris chacei Williams and Rona 1986: a possible symbiosis. PLoS One 13:e0206084. 10.1371/journal.pone.0206084 - DOI - PMC - PubMed
-
- Bergh Ø. (1995). Bacteria associated with early life stages of halibut, Hippoglossus hippoglossus L., inhibit growth of a pathogenic Vibrio sp. J. Fish Dis. 18 31–40. 10.1111/j.1365-2761.1995.tb01263.x - DOI
-
- Breiman L. (2001). Random forests. Mach. Learn. 45 5–32. 10.1023/A:1010933404324 - DOI
LinkOut - more resources
Full Text Sources

