Analysis of Breast Cancer Based on the Dysregulated Network
- PMID: 35242172
- PMCID: PMC8886234
- DOI: 10.3389/fgene.2022.856075
Analysis of Breast Cancer Based on the Dysregulated Network
Abstract
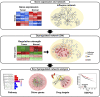
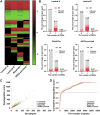
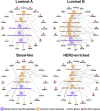
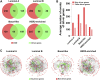
Breast cancer is a heterogeneous disease, and its development is closely associated with the underlying molecular regulatory network. In this paper, we propose a new way to measure the regulation strength between genes based on their expression values, and construct the dysregulated networks (DNs) for the four subtypes of breast cancer. Our results show that the key dysregulated networks (KDNs) are significantly enriched in critical breast cancer-related pathways and driver genes; closely related to drug targets; and have significant differences in survival analysis. Moreover, the key dysregulated genes could serve as potential driver genes, drug targets, and prognostic markers for each breast cancer subtype. Therefore, the KDN is expected to be an effective and novel way to understand the mechanisms of breast cancer.
Keywords: breast cancer; cancer-related pathways; driver genes; drug targets; dysregulated network; survival analysis.
Copyright © 2022 Huo, Li, Xu, Bao and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Immunoglobulin superfamily genes are novel prognostic biomarkers for breast cancer.Oncotarget. 2017 Jan 10;8(2):2444-2456. doi: 10.18632/oncotarget.13683. Oncotarget. 2017. PMID: 27911271 Free PMC article.
-
Comprehensive Analysis of Differentially Expressed Profiles of lncRNAs/mRNAs and miRNAs with Associated ceRNA Networks in Triple-Negative Breast Cancer.Cell Physiol Biochem. 2018;50(2):473-488. doi: 10.1159/000494162. Epub 2018 Oct 11. Cell Physiol Biochem. 2018. PMID: 30308479
-
Network-based method for detecting dysregulated pathways in glioblastoma cancer.IET Syst Biol. 2018 Feb;12(1):39-44. doi: 10.1049/iet-syb.2017.0033. IET Syst Biol. 2018. PMID: 29337288 Free PMC article.
-
Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics.Onco Targets Ther. 2016 Feb 12;9:761-72. doi: 10.2147/OTT.S97664. eCollection 2016. Onco Targets Ther. 2016. PMID: 26929647 Free PMC article.
-
Identifying driver genes involving gene dysregulated expression, tissue-specific expression and gene-gene network.BMC Med Genomics. 2019 Dec 30;12(Suppl 7):168. doi: 10.1186/s12920-019-0619-z. BMC Med Genomics. 2019. PMID: 31888619 Free PMC article.
Cited by
-
Patterns of Immunohistochemical Expression of P53, BCL2, PTEN, and HER2/neu Tumor Markers in Specific Breast Cancer Lesions.Evid Based Complement Alternat Med. 2022 Oct 17;2022:2026284. doi: 10.1155/2022/2026284. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 36299777 Free PMC article.
-
Cellular and Exosomal MicroRNAs: Emerging Clinical Relevance as Targets for Breast Cancer Diagnosis and Prognosis.Adv Biol (Weinh). 2024 Apr;8(4):e2300532. doi: 10.1002/adbi.202300532. Epub 2024 Jan 22. Adv Biol (Weinh). 2024. PMID: 38258348 Free PMC article. Review.
-
Integrative network analysis of transcriptomics data reveals potential prognostic biomarkers for colorectal cancer.Cancer Med. 2024 Jun;13(11):e7391. doi: 10.1002/cam4.7391. Cancer Med. 2024. PMID: 38872418 Free PMC article.
References
-
- Bao Z., Zhu Y., Ge Q., Gu W., Bai Y. J. I. A. (2020). Signaling Pathway Analysis Combined with the Strength Variations of Interactions between Genes under Different Conditions, 1.
LinkOut - more resources
Full Text Sources

