Expression signature of the Leigh syndrome French-Canadian type
- PMID: 35242578
- PMCID: PMC8856909
- DOI: 10.1016/j.ymgmr.2022.100847
Expression signature of the Leigh syndrome French-Canadian type
Abstract
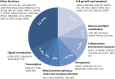
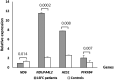
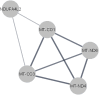
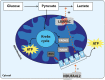
As a result of a founder effect, a Leigh syndrome variant called Leigh syndrome, French-Canadian type (LSFC, MIM / 220,111) is more frequent in Saguenay-Lac-Saint-Jean (SLSJ), a geographically isolated region on northeastern Quebec, Canada. LSFC is a rare autosomal recessive mitochondrial neurodegenerative disorder due to damage in mitochondrial energy production. LSFC is caused by pathogenic variants in the nuclear gene leucine-rich pentatricopeptide repeat-containing (LRPPRC). Despite progress understanding the molecular mode of action of LRPPRC gene, there is no treatment for this disease. The present study aims to identify the biological pathways altered in the LSFC disorder through microarray-based transcriptomic profile analysis of twelve LSFC cell lines compared to twelve healthy ones, followed by gene ontology (GO) and pathway analyses. A set of 84 significantly differentially expressed genes were obtained (p ≥ 0.05; Fold change (Flc) ≥ 1.5). 45 genes were more expressed (53.57%) in LSFC cell lines compared to controls and 39 (46.43%) had lower expression levels. Gene ontology analysis highlighted altered expression of genes involved in the mitochondrial respiratory chain and energy production, glucose and lipids metabolism, oncogenesis, inflammation and immune response, cell growth and apoptosis, transcription, and signal transduction. Considering the metabolic nature of LSFC disease, genes included in the mitochondrial respiratory chain and energy production cluster stood out as the most important ones to be involved in LSFC mitochondrial disorder. In addition, the protein-protein interaction network indicated a strong interaction between the genes included in this cluster. The mitochondrial gene NDUFA4L2 (NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, 4-like 2), with higher expression in LSFC cells, represents a target for functional studies to explain the role of this gene in LSFC disease. This work provides, for the first time, the LSFC gene expression profile in fibroblasts isolated from affected individuals. This represents a valuable resource to understand the pathogenic basis and consequences of LRPPRC dysfunction.
Keywords: ATP, adénosine-5'-triphosphate; COPD, chronic obstructive pulmonary disease; COX, cytochrome c-oxidase; Cytochrome c oxidase; DMEM, Dubelcco’s Modified Essential Medium; ETC, electron transport chain; Flc, fold change; GO, gene ontology; Gene expression; HES1, hairy and enhancer of split 1; HIF-1, hypoxia inducible factor-1; LRPPRC; LRPPRC, leucine-rich pentatricopeptide repeat-containing; LSFC, Leigh syndrome, French-Canadian type; Leigh syndrome; Leigh syndrome French-Canadian type (LSFC); Microarrays; Mitochondrial chain respiration; NAFLD, non-alcoholic fatty liver disease; ND6, NADH dehydrogenase, subunit 6; NDUFA4L2; NDUFA4L2, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, 4-like 2; OXPHOS, oxidative phosphorylation; PFKFB4, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4; PPI, protein‐protein interaction; RMA, robust multi-array analysis; ROS, reactive oxygen species; RPL13A, ribosomal protein L13a; SLIRP, stem-loop interacting protein; SLSJ, Saguenay–Lac-Saint-Jean; SRA, steroid receptor RNA activator; qRT-PCR, Real-time PCR; rare diseases.
© 2022 The Authors.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials

