A plant receptor domain with functional analogies to animal malectin disables ER stress responses upon infection
- PMID: 35243239
- PMCID: PMC8861646
- DOI: 10.1016/j.isci.2022.103877
A plant receptor domain with functional analogies to animal malectin disables ER stress responses upon infection
Abstract
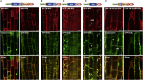
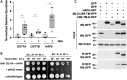
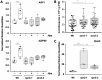
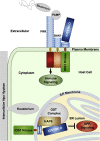
Malectins from the oligosaccharyltransferase (OST) complex in the endoplasmic reticulum (ER) of animal cells are involved in ER quality control and contribute to the Unfolded Protein Response (UPR). Malectins are not found in plant cells, but malectin-like domains (MLDs) are constituents of many membrane-bound receptors. In Arabidopsis thaliana, the MLD-containing receptor IOS1 promotes successful infection by filamentous plant pathogens. We show that the MLD of its exodomain retains IOS1 in the ER of plant cells and attenuates the infection-induced UPR. Expression of the MLD in the ios1-1 knockout background is sufficient to complement infection-related phenotypes of the mutant, such as increased UPR and reduced disease susceptibility. IOS1 interacts with the ER membrane-associated ribophorin HAP6 from the OST complex, and hap6 mutants show decreased pathogen-responsive UPR and increased disease susceptibility. Altogether, this study revealed a previously uncharacterized role of a plant receptor domain in the regulation of ER stress during infection.
Keywords: Molecular biology; Plant biology; Plant pathology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bittner-Eddy P.D., Allen R.L., Rehmany A.P., Birch P., Beynon J.L. Use of suppression subtractive hybridization to identify downy mildew genes expressed during infection of Arabidopsis thaliana. Mol. Plant Pathol. 2003;4:501–507. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

