Auto-CORPus: A Natural Language Processing Tool for Standardizing and Reusing Biomedical Literature
- PMID: 35243479
- PMCID: PMC8885717
- DOI: 10.3389/fdgth.2022.788124
Auto-CORPus: A Natural Language Processing Tool for Standardizing and Reusing Biomedical Literature
Abstract
To analyse large corpora using machine learning and other Natural Language Processing (NLP) algorithms, the corpora need to be standardized. The BioC format is a community-driven simple data structure for sharing text and annotations, however there is limited access to biomedical literature in BioC format and a lack of bioinformatics tools to convert online publication HTML formats to BioC. We present Auto-CORPus (Automated pipeline for Consistent Outputs from Research Publications), a novel NLP tool for the standardization and conversion of publication HTML and table image files to three convenient machine-interpretable outputs to support biomedical text analytics. Firstly, Auto-CORPus can be configured to convert HTML from various publication sources to BioC. To standardize the description of heterogenous publication sections, the Information Artifact Ontology is used to annotate each section within the BioC output. Secondly, Auto-CORPus transforms publication tables to a JSON format to store, exchange and annotate table data between text analytics systems. The BioC specification does not include a data structure for representing publication table data, so we present a JSON format for sharing table content and metadata. Inline tables within full-text HTML files and linked tables within separate HTML files are processed and converted to machine-interpretable table JSON format. Finally, Auto-CORPus extracts abbreviations declared within publication text and provides an abbreviations JSON output that relates an abbreviation with the full definition. This abbreviation collection supports text mining tasks such as named entity recognition by including abbreviations unique to individual publications that are not contained within standard bio-ontologies and dictionaries. The Auto-CORPus package is freely available with detailed instructions from GitHub at: https://github.com/omicsNLP/Auto-CORPus.
Keywords: biomedical literature; health data; natural language processing; semantics; text mining.
Copyright © 2022 Beck, Shorter, Hu, Li, Sun, Popovici, McQuibban, Makraduli, Yeung, Rowlands and Posma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


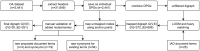


References
-
- Jackson RG, Patel R, Jayatilleke N, Kolliakou A, Ball M, Gorrell G, et al. Natural language processing to extract symptoms of severe mental illness from clinical text: the Clinical Record Interactive Search Comprehensive Data Extraction (CRIS-CODE) project. BMJ Open. (2017) 7:e012012. 10.1136/bmjopen-2016-012012 - DOI - PMC - PubMed
-
- Wang LL, Cachola I, Bragg J, Yu-Yen Cheng E, Haupt C, Latzke M, et al. Improving the accessibility of scientific documents: current state, user needs, and a system solution to enhance scientific PDF accessibility for blind and low vision users. arXiv e-prints: arXiv:2105.00076 (2021). Available online at: https://arxiv.org/pdf/2105.00076.pdf
Grants and funding
LinkOut - more resources
Full Text Sources

