First cases of infection with the 21L/BA.2 Omicron variant in Marseille, France
- PMID: 35243660
- PMCID: PMC9088623
- DOI: 10.1002/jmv.27695
First cases of infection with the 21L/BA.2 Omicron variant in Marseille, France
Abstract
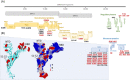
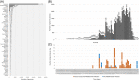
The SARS-CoV-2 21K/BA.1, 21L/BA.2, and BA.3 Omicron variants have recently emerged worldwide. To date, the 21L/BA.2 Omicron variant has remained very minority globally but became predominant in Denmark instead of the 21K/BA.1 variant. Here, we describe the first cases diagnosed with this variant in south-eastern France. We identified 13 cases using variant-specific qPCR and next-generation sequencing between 28/11/2021 and 31/01/2022, the first two cases being diagnosed in travelers returning from Tanzania. Overall, viral genomes displayed a mean (±standard deviation) number of 65.9 ± 2.5 (range, 61-69) nucleotide substitutions and 31.0 ± 8.3 (27-50) nucleotide deletions, resulting in 49.6 ± 2.2 (45-52) amino acid substitutions (including 28 in the spike protein) and 12.4 ± 1.1 (12-15) amino acid deletions. Phylogeny showed the distribution in three different clusters of these genomes, which were most closely related to genomes from England and South Africa, from Singapore and Nepal, or from France and Denmark. Structural predictions highlighted a significant enlargement and flattening of the surface of the 21L/BA.2 N-terminal domain of the spike protein compared to that of the 21K/BA.1 Omicron variant, which may facilitate initial viral interactions with lipid rafts. Close surveillance is needed at global, country, and center scales to monitor the incidence and clinical outcome of the 21L/BA.2 Omicron variant.
Keywords: Omicron; SARS-CoV-2; emergence; southern France; travel; variant.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
All authors have no conflicts of interest to declare. Didier Raoult has been a consultant for Hitachi High‐Technologies Corporation, Tokyo, Japan from 2018 to 2020. He is a scientific board member of Eurofins company and a founder of a microbial culture company (Culture Top).
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

