Privacy-preserving quality control of neuroimaging datasets in federated environments
- PMID: 35243723
- PMCID: PMC8996357
- DOI: 10.1002/hbm.25788
Privacy-preserving quality control of neuroimaging datasets in federated environments
Abstract
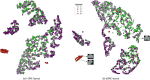
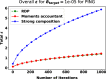
Privacy concerns for rare disease data, institutional or IRB policies, access to local computational or storage resources or download capabilities are among the reasons that may preclude analyses that pool data to a single site. A growing number of multisite projects and consortia were formed to function in the federated environment to conduct productive research under constraints of this kind. In this scenario, a quality control tool that visualizes decentralized data in its entirety via global aggregation of local computations is especially important, as it would allow the screening of samples that cannot be jointly evaluated otherwise. To solve this issue, we present two algorithms: decentralized data stochastic neighbor embedding, dSNE, and its differentially private counterpart, DP-dSNE. We leverage publicly available datasets to simultaneously map data samples located at different sites according to their similarities. Even though the data never leaves the individual sites, dSNE does not provide any formal privacy guarantees. To overcome that, we rely on differential privacy: a formal mathematical guarantee that protects individuals from being identified as contributors to a dataset. We implement DP-dSNE with AdaCliP, a method recently proposed to add less noise to the gradients per iteration. We introduce metrics for measuring the embedding quality and validate our algorithms on these metrics against their centralized counterpart on two toy datasets. Our validation on six multisite neuroimaging datasets shows promising results for the quality control tasks of visualization and outlier detection, highlighting the potential of our private, decentralized visualization approach.
Keywords: fMRI; federated neuroimaging; quality control; sMRI.
© 2022 The Authors. Human Brain Mapping published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures






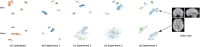





References
-
- Amir, G. , Gal, C. , Fernando, P. , & Naftali, T. (2007). Euclidean embedding of co‐occurrence data. Journal of Machine Learning Research, 8, 2265–2295.
-
- Baker, B. T. , Silva, R. , Calhoun, V. , Sarwate, A. , & Plis, S. (2015). Large scale collaboration with autonomy: Decentralized data ICA. In 2015 IEEE 25th International Workshop on Machine Learning for Signal Processing (MLSP) (pp. 1–6). Boston, MA: IEEE.
-
- Brendan McMahan H., Eider Moore, Daniel Ramage, Agüera Arcas Blaise. (2016) Federated learning of deep networks using model averaging. CoRR.2016;abs/1602.05629.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

