Assembly of recombinant tau into filaments identical to those of Alzheimer's disease and chronic traumatic encephalopathy
- PMID: 35244536
- PMCID: PMC8983045
- DOI: 10.7554/eLife.76494
Assembly of recombinant tau into filaments identical to those of Alzheimer's disease and chronic traumatic encephalopathy
Abstract
Abundant filamentous inclusions of tau are characteristic of more than 20 neurodegenerative diseases that are collectively termed tauopathies. Electron cryo-microscopy (cryo-EM) structures of tau amyloid filaments from human brain revealed that distinct tau folds characterise many different diseases. A lack of laboratory-based model systems to generate these structures has hampered efforts to uncover the molecular mechanisms that underlie tauopathies. Here, we report in vitro assembly conditions with recombinant tau that replicate the structures of filaments from both Alzheimer's disease (AD) and chronic traumatic encephalopathy (CTE), as determined by cryo-EM. Our results suggest that post-translational modifications of tau modulate filament assembly, and that previously observed additional densities in AD and CTE filaments may arise from the presence of inorganic salts, like phosphates and sodium chloride. In vitro assembly of tau into disease-relevant filaments will facilitate studies to determine their roles in different diseases, as well as the development of compounds that specifically bind to these structures or prevent their formation.
Keywords: E. coli; amyloids; cryo-EM; human; molecular biophysics; neuroscience; structural biology; tauopathies.
Plain language summary
Many neurodegenerative diseases, including Alzheimer’s disease, the most common form of dementia, are characterised by knotted clumps of a protein called tau. In these diseases, tau misfolds, stacks together and forms abnormal filaments, which have a structured core and fuzzy coat. These sticky, misfolded proteins are thought to be toxic to brain cells, the loss of which ultimately causes problems with how people move, think, feel or behave. Reconstructing the shape of tau filaments using an atomic-level imaging technique called electron cryo-microscopy, or cryo-EM, researchers have found distinct types of tau filaments present in certain diseases. In Alzheimer’s disease, for example, a mixture of paired helical and straight filaments is found. Different tau filaments are seen again in chronic traumatic encephalopathy (CTE), a condition associated with repetitive brain trauma. It remains unclear, however, how tau folds into these distinct shapes and under what conditions it forms certain types of filaments. The role that distinct tau folds play in different diseases is also poorly understood. This is largely because researchers making tau proteins in the lab have yet to replicate the exact structure of tau filaments found in diseased brain tissue. Lövestam et al. describe the conditions for making tau filaments in the lab identical to those isolated from the brains of people who died from Alzheimer’s disease and CTE. Lövestam et al. instructed bacteria to make tau protein, optimised filament assembly conditions, including shaking time and speed, and found that bona fide filaments formed from shortened versions of tau. On cryo-EM imaging, the lab-produced filaments had the same left-handed twist and helical symmetry as filaments characteristic of Alzheimer’s disease. Adding salts, however, changed the shape of tau filaments. In the presence of sodium chloride, otherwise known as kitchen salt, tau formed filaments with a filled cavity at the core, identical to tau filaments observed in CTE. Again, this structure was confirmed on cryo-EM imaging. Being able to make tau filaments identical to those found in human tauopathies will allow scientists to study how these filaments form and elucidate what role they play in disease. Ultimately, a better understanding of tau filament formation could lead to improved diagnostics and treatments for neurodegenerative diseases involving tau.
© 2022, Lövestam et al.
Conflict of interest statement
SL, AM, MG No competing interests declared, FK, Bv, AK is affiliated with Thermo Fisher Scientific. The author has no financial interests to declare, SS Reviewing editor, eLife
Figures







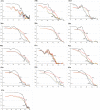
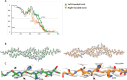



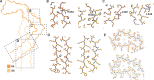


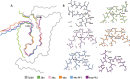
References
-
- Al-Hilaly YK, Pollack SJ, Vadukul DM, Citossi F, Rickard JE, Simpson M, Storey JMD, Harrington CR, Wischik CM, Serpell LC. Alzheimer’s Disease-like Paired Helical Filament Assembly from Truncated Tau Protein Is Independent of Disulfide Crosslinking. Journal of Molecular Biology. 2017;429:3650–3665. doi: 10.1016/j.jmb.2017.09.007. - DOI - PubMed
-
- Al-Hilaly YK, Foster BE, Biasetti L, Lutter L, Pollack SJ, Rickard JE, Storey JMD, Harrington CR, Xue WF, Wischik CM, Serpell LC. Tau (297-391) forms filaments that structurally mimic the core of paired helical filaments in Alzheimer’s disease brain. FEBS Letters. 2020;594:944–950. doi: 10.1002/1873-3468.13675. - DOI - PMC - PubMed
-
- Arakhamia T, Lee CE, Carlomagno Y, Kumar M, Duong DM, Wesseling H, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, Seyfried NT, Petrucelli L, Steen JA, Fitzpatrick AWP. Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell. 2021;184:6207–6210. doi: 10.1016/j.cell.2021.11.029. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

