Structural dynamics determine voltage and pH gating in human voltage-gated proton channel
- PMID: 35244539
- PMCID: PMC8926398
- DOI: 10.7554/eLife.73093
Structural dynamics determine voltage and pH gating in human voltage-gated proton channel
Abstract
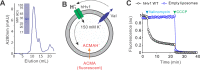
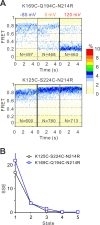
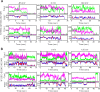
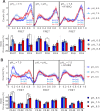
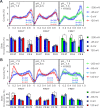
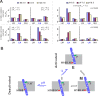
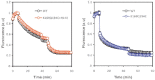
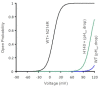
Voltage-gated proton (Hv) channels are standalone voltage sensors without separate ion conductive pores. They are gated by both voltage and transmembrane proton gradient (i.e., ∆pH), serving as acid extruders in most cells. Like the canonical voltage sensors, Hv channels are a bundle of four helices (named S1 -S4), with the S4 segment carrying three positively charged Arg residues. Extensive structural and electrophysiological studies on voltage-gated ion channels, in general, agree on an outwards movement of the S4 segment upon activating voltage, but the real-time conformational transitions are still unattainable. With purified human voltage-gated proton (hHv1) channels reconstituted in liposomes, we have examined its conformational dynamics, including the S4 segment at different voltage and pHs using single-molecule fluorescence resonance energy transfer (smFRET). Here, we provide the first glimpse of real-time conformational trajectories of the hHv1 voltage sensor and show that both voltage and pH gradient shift the conformational dynamics of the S4 segment to control channel gating. Our results indicate that the S4 segment transits among three major conformational states and only the transitions between the inward and outward conformations are highly dependent on voltage and pH. Altogether, we propose a kinetic model that explains the mechanisms underlying voltage and pH gating in Hv channels, which may also serve as a general framework for understanding the voltage sensing and gating in other voltage-gated ion channels.
Keywords: E. coli; human; ion channel; membrane protein; molecular biophysics; proton channel; single-molecule FRET; structural biology; structural dynamics; voltage gating.
© 2022, Han et al.
Conflict of interest statement
SH, SP, JV, KT, ND, NB, ZG, SW No competing interests declared
Figures




Similar articles
-
Cholesterol inhibits human voltage-gated proton channel hHv1.Proc Natl Acad Sci U S A. 2022 Sep 6;119(36):e2205420119. doi: 10.1073/pnas.2205420119. Epub 2022 Aug 29. Proc Natl Acad Sci U S A. 2022. PMID: 36037383 Free PMC article.
-
Arachidonic acid reverses cholesterol and zinc inhibition of human voltage-gated proton channels.J Biol Chem. 2023 Jul;299(7):104918. doi: 10.1016/j.jbc.2023.104918. Epub 2023 Jun 12. J Biol Chem. 2023. PMID: 37315791 Free PMC article.
-
Voltage sensors.Mol Pharmacol. 2025 Feb;107(2):100011. doi: 10.1016/j.molpha.2024.100011. Epub 2024 Dec 12. Mol Pharmacol. 2025. PMID: 40023511 Review.
-
Voltage sensor dynamics of a bacterial voltage-gated sodium channel NavAb reveal three conformational states.J Biol Chem. 2023 Mar;299(3):102967. doi: 10.1016/j.jbc.2023.102967. Epub 2023 Feb 1. J Biol Chem. 2023. PMID: 36736429 Free PMC article.
-
Emerging issues of connexin channels: biophysics fills the gap.Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
Cited by
-
Cholesterol inhibits human voltage-gated proton channel hHv1.Proc Natl Acad Sci U S A. 2022 Sep 6;119(36):e2205420119. doi: 10.1073/pnas.2205420119. Epub 2022 Aug 29. Proc Natl Acad Sci U S A. 2022. PMID: 36037383 Free PMC article.
-
A novel method for expressing and purifying large quantities of functional and stable human voltage-gated proton channel (hHv1).Protein Sci. 2025 Feb;34(2):e70017. doi: 10.1002/pro.70017. Protein Sci. 2025. PMID: 39865375
-
Expression and Purification of the Human Voltage-Gated Proton Channel (hHv1).Bio Protoc. 2025 Jun 20;15(12):e5344. doi: 10.21769/BioProtoc.5344. eCollection 2025 Jun 20. Bio Protoc. 2025. PMID: 40657542 Free PMC article.
-
Voltage sensor conformations induced by LQTS-associated mutations in hERG potassium channels.Nat Commun. 2025 Aug 3;16(1):7126. doi: 10.1038/s41467-025-62472-9. Nat Commun. 2025. PMID: 40753176 Free PMC article.
-
Voltage-Gated Proton Channels in the Tree of Life.Biomolecules. 2023 Jun 24;13(7):1035. doi: 10.3390/biom13071035. Biomolecules. 2023. PMID: 37509071 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

