CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells
- PMID: 35244715
- PMCID: PMC9226543
- DOI: 10.1093/nar/gkac159
CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells
Abstract
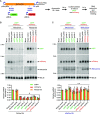
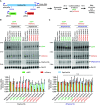
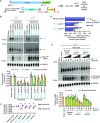
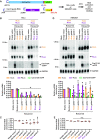
CRISPR/Cas13 effectors have garnered increasing attention as easily customizable tools for detecting and depleting RNAs of interest. Near perfect complementarity between a target RNA and the Cas13-associated guide RNA is required for activation of Cas13 ribonuclease activity. Nonetheless, the specificity of Cas13 effectors in eukaryotic cells has been debated as the Cas13 nuclease domains can be exposed on the enzyme surface, providing the potential for promiscuous cleavage of nearby RNAs (so-called collateral damage). Here, using co-transfection assays in Drosophila and human cells, we found that the off-target effects of RxCas13d, a commonly used Cas13 effector, can be as strong as the level of on-target RNA knockdown. The extent of off-target effects is positively correlated with target RNA expression levels, and collateral damage can be observed even after reducing RxCas13d/guide RNA levels. The PspCas13b effector showed improved specificity and, unlike RxCas13d, can be used to deplete a Drosophila circular RNA without affecting the expression of the associated linear RNA. PspCas13b nonetheless still can have off-target effects and we notably found that the extent of off-target effects for Cas13 effectors differs depending on the cell type and target RNA examined. In total, these results highlight the need for caution when designing and interpreting Cas13-based knockdown experiments.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

