Selective requirement for polycomb repressor complex 2 in the generation of specific hypothalamic neuronal subtypes
- PMID: 35245348
- PMCID: PMC8959139
- DOI: 10.1242/dev.200076
Selective requirement for polycomb repressor complex 2 in the generation of specific hypothalamic neuronal subtypes
Abstract
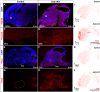
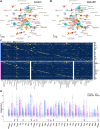
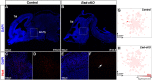
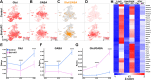
The hypothalamus displays staggering cellular diversity, chiefly established during embryogenesis by the interplay of several signalling pathways and a battery of transcription factors. However, the contribution of epigenetic cues to hypothalamus development remains unclear. We mutated the polycomb repressor complex 2 gene Eed in the developing mouse hypothalamus, which resulted in the loss of H3K27me3, a fundamental epigenetic repressor mark. This triggered ectopic expression of posteriorly expressed regulators (e.g. Hox homeotic genes), upregulation of cell cycle inhibitors and reduced proliferation. Surprisingly, despite these effects, single cell transcriptomic analysis revealed that most neuronal subtypes were still generated in Eed mutants. However, we observed an increase in glutamatergic/GABAergic double-positive cells, as well as loss/reduction of dopamine, hypocretin and Tac2-Pax6 neurons. These findings indicate that many aspects of the hypothalamic gene regulatory flow can proceed without the key H3K27me3 epigenetic repressor mark, but points to a unique sensitivity of particular neuronal subtypes to a disrupted epigenomic landscape.
Keywords: Cell specification; Epigenetics; H3K27me3; H3K4me1/3; Mouse; Neuropeptide neurons.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures