Genome-wide association study reveals novel genetic loci: a new polygenic risk score for mitral valve prolapse
- PMID: 35245370
- PMCID: PMC9649914
- DOI: 10.1093/eurheartj/ehac049
Genome-wide association study reveals novel genetic loci: a new polygenic risk score for mitral valve prolapse
Abstract
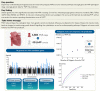
Aims: Mitral valve prolapse (MVP) is a common valvular heart disease with a prevalence of >2% in the general adult population. Despite this high incidence, there is a limited understanding of the molecular mechanism of this disease, and no medical therapy is available for this disease. We aimed to elucidate the genetic basis of MVP in order to better understand this complex disorder.
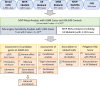
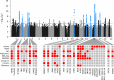
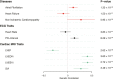
Methods and results: We performed a meta-analysis of six genome-wide association studies that included 4884 cases and 434 649 controls. We identified 14 loci associated with MVP in our primary analysis and 2 additional loci associated with a subset of the samples that additionally underwent mitral valve surgery. Integration of epigenetic, transcriptional, and proteomic data identified candidate MVP genes including LMCD1, SPTBN1, LTBP2, TGFB2, NMB, and ALPK3. We created a polygenic risk score (PRS) for MVP and showed an improved MVP risk prediction beyond age, sex, and clinical risk factors.
Conclusion: We identified 14 genetic loci that are associated with MVP. Multiple analyses identified candidate genes including two transforming growth factor-β signalling molecules and spectrin β. We present the first PRS for MVP that could eventually aid risk stratification of patients for MVP screening in a clinical setting. These findings advance our understanding of this common valvular heart disease and may reveal novel therapeutic targets for intervention.
Keywords: Genetic correlation; Genome-wide association study; Mitral valve prolapse; Polygenic risk score; Proteomics; RNA-sequencing.
© The Author(s) 2022. Published by Oxford University Press on behalf of European Society of Cardiology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Comment in
-
Mitral valve prolapse: will genetics finally solve the puzzle?Eur Heart J. 2022 May 1;43(17):1681-1683. doi: 10.1093/eurheartj/ehac048. Eur Heart J. 2022. PMID: 35265976 No abstract available.
References
-
- Freed LA, Levy D, Levine RA, Larson MG, Evans JC, Fuller DL, et al. Prevalence and clinical outcome of mitral-valve prolapse. N Engl J Med 1999;341:1–7. - PubMed
-
- Vahanian A, Beyersdorf F, Praz F, Milojevic M, Baldus S, Bauersachs J, et al. 2021 ESC/EACTS Guidelines for the management of valvular heart disease: developed by the task force for the management of valvular heart disease of the European Society of Cardiology (ESC) and the European Association for Cardio-Thoracic Surgery (EACTS). Eur Heart J 2021;43:561–632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

