Kynurenine importation by SLC7A11 propagates anti-ferroptotic signaling
- PMID: 35245456
- PMCID: PMC7617904
- DOI: 10.1016/j.molcel.2022.02.007
Kynurenine importation by SLC7A11 propagates anti-ferroptotic signaling
Abstract
IDO1 oxidizes tryptophan (TRP) to generate kynurenine (KYN), the substrate for 1-carbon and NAD metabolism, and is implicated in pro-cancer pathophysiology and infection biology. However, the mechanistic relationships between IDO1 in amino acid depletion versus product generation have remained a longstanding mystery. We found an unrecognized link between IDO1 and cell survival mediated by KYN that serves as the source for molecules that inhibit ferroptotic cell death. We show that this effect requires KYN export from IDO1-expressing cells, which is then available for non-IDO1-expressing cells via SLC7A11, the central transporter involved in ferroptosis suppression. Whether inside the "producer" IDO1+ cell or the "receiver" cell, KYN is converted into downstream metabolites, suppressing ferroptosis by ROS scavenging and activating an NRF2-dependent, AHR-independent cell-protective pathway, including SLC7A11, propagating anti-ferroptotic signaling. IDO1, therefore, controls a multi-pronged protection pathway from ferroptotic cell death, underscoring the need to re-evaluate the use of IDO1 inhibitors in cancer treatment.
Keywords: GCN2; IDO1; NRF2; SLC7A11; cancer; ferroptosis; kynurenine; metabolism; tryptophan.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests directly related to this work. Thomas Köcher is affiliated with Vienna BioCenter Core Facilities GmbH. Peter J. Murray is on the scientific advisory boards of Palleon Pharma and ImCheck Pharm, which is unrelated to the content herein.
Figures

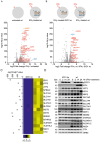



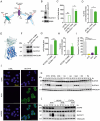
References
-
- Arensman MD, Yang XS, Leahy DM, Toral-Barza L, Mileski M, Rosfjord EC, Wang F, Deng S, Myers JS, Abraham RT, et al. Cystine-glutamate antiporter xCT deficiency suppresses tumor growth while preserving antitumor immunity. Proc Natl Acad Sci USA. 2019;116:9533–9542. doi: 10.1073/pnas.1814932116. - DOI - PMC - PubMed
-
- Austin CJ, Mailu BM, Maghzal GJ, Sanchez-Perez A, Rahlfs S, Zocher K, Yuasa HJ, Arthur JW, Becker K, Stocker R, et al. Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2. Amino Acids. 2010;39:565–578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

