Host-microbiome protein-protein interactions capture disease-relevant pathways
- PMID: 35246229
- PMCID: PMC8895870
- DOI: 10.1186/s13059-022-02643-9
Host-microbiome protein-protein interactions capture disease-relevant pathways
Abstract
Background: Host-microbe interactions are crucial for normal physiological and immune system development and are implicated in a variety of diseases, including inflammatory bowel disease (IBD), colorectal cancer (CRC), obesity, and type 2 diabetes (T2D). Despite large-scale case-control studies aimed at identifying microbial taxa or genes involved in pathogeneses, the mechanisms linking them to disease have thus far remained elusive.
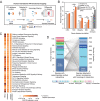
Results: To identify potential pathways through which human-associated bacteria impact host health, we leverage publicly-available interspecies protein-protein interaction (PPI) data to find clusters of microbiome-derived proteins with high sequence identity to known human-protein interactors. We observe differential targeting of putative human-interacting bacterial genes in nine independent metagenomic studies, finding evidence that the microbiome broadly targets human proteins involved in immune, oncogenic, apoptotic, and endocrine signaling pathways in relation to IBD, CRC, obesity, and T2D diagnoses.
Conclusions: This host-centric analysis provides a mechanistic hypothesis-generating platform and extensively adds human functional annotation to commensal bacterial proteins.
Keywords: Gut microbiome; Human disease; Metagenomics; Protein-protein interactions.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Fungal microbiome in inflammatory bowel disease: a critical assessment.J Clin Invest. 2022 Mar 1;132(5):e155786. doi: 10.1172/JCI155786. J Clin Invest. 2022. PMID: 35229726 Free PMC article. Review.
-
Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health.Gut Microbes. 2020 Sep 2;11(5):1300-1313. doi: 10.1080/19490976.2020.1748261. Epub 2020 Apr 24. Gut Microbes. 2020. PMID: 32329665 Free PMC article.
-
Clustering co-abundant genes identifies components of the gut microbiome that are reproducibly associated with colorectal cancer and inflammatory bowel disease.Microbiome. 2019 Aug 1;7(1):110. doi: 10.1186/s40168-019-0722-6. Microbiome. 2019. PMID: 31370880 Free PMC article.
-
Microbial genes and pathways in inflammatory bowel disease.Nat Rev Microbiol. 2019 Aug;17(8):497-511. doi: 10.1038/s41579-019-0213-6. Nat Rev Microbiol. 2019. PMID: 31249397 Free PMC article. Review.
-
Gene-level metagenomic architectures across diseases yield high-resolution microbiome diagnostic indicators.Nat Commun. 2021 May 18;12(1):2907. doi: 10.1038/s41467-021-23029-8. Nat Commun. 2021. PMID: 34006865 Free PMC article.
Cited by
-
Choosing Therapies in Ulcerative Colitis.J Can Assoc Gastroenterol. 2023 Sep 4;7(1):9-21. doi: 10.1093/jcag/gwad025. eCollection 2024 Feb. J Can Assoc Gastroenterol. 2023. PMID: 38314181 Free PMC article.
-
HMI-PRED 2.0: a biologist-oriented web application for prediction of host-microbe protein-protein interaction by interface mimicry.Bioinformatics. 2022 Oct 31;38(21):4962-4965. doi: 10.1093/bioinformatics/btac633. Bioinformatics. 2022. PMID: 36124958 Free PMC article.
-
Exploring the impact of pathogenic microbiome in orthopedic diseases: machine learning and deep learning approaches.Front Cell Infect Microbiol. 2024 Apr 3;14:1380136. doi: 10.3389/fcimb.2024.1380136. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38633744 Free PMC article. Review.
-
RIscoper 2.0: A deep learning tool to extract RNA biomedical relation sentences from literature.Comput Struct Biotechnol J. 2024 Mar 24;23:1469-1476. doi: 10.1016/j.csbj.2024.03.017. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38623560 Free PMC article.
-
Proteomics and Microbiota Conjoint Analysis in the Nasal Mucus: Revelation of Differences in Immunological Function in Manis javanica and Manis pentadactyla.Animals (Basel). 2024 Sep 14;14(18):2683. doi: 10.3390/ani14182683. Animals (Basel). 2024. PMID: 39335272 Free PMC article.
References
-
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834–841. - PubMed
-
- Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107–113. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

