The space between notes: emerging roles for translationally silent ribosomes
- PMID: 35246374
- PMCID: PMC9106873
- DOI: 10.1016/j.tibs.2022.02.003
The space between notes: emerging roles for translationally silent ribosomes
Abstract
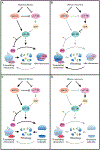
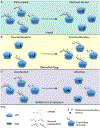
In addition to their central functions in translation, ribosomes can adopt inactive structures that are fully assembled yet devoid of mRNA. We describe how the abundance of idle eukaryotic ribosomes is influenced by a broad range of biological conditions spanning viral infection, nutrient deprivation, and developmental cues. Vacant ribosomes may provide a means to exclude ribosomes from translation while also shielding them from degradation, and the variable identity of factors that occlude ribosomes may impart distinct functionality. We propose that regulated changes in the balance of idle and active ribosomes provides a means to fine-tune translation. We provide an overview of idle ribosomes, describe what is known regarding their function, and highlight questions that may clarify their biological roles.
Keywords: eEF2K; hibernating ribosome; idle ribosome; mTOR; vacant ribosome.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests No interests are declared.
Figures

References
-
- Prossliner T et al. (2018) Ribosome Hibernation. Annu. Rev. Genet 52, 321–348 - PubMed
-
- Anger AM et al. (2013) Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 - PubMed
-
- Ben-Shem A et al. (2011) The Structure of the Eukaryotic Ribosome at 3.0 Å Resolution. Science (80-.) 334, 1524 LP–1529 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

