In PD-1+ human colon cancer cells NIVOLUMAB promotes survival and could protect tumor cells from conventional therapies
- PMID: 35246475
- PMCID: PMC8900051
- DOI: 10.1136/jitc-2021-004032
In PD-1+ human colon cancer cells NIVOLUMAB promotes survival and could protect tumor cells from conventional therapies
Abstract
Background: Colorectal cancer (CRC) is one of the most prevalent and deadly tumors worldwide. The majority of CRC is resistant to anti-programmed cell death-1 (PD-1)-based cancer immunotherapy, with approximately 15% with high-microsatellite instability, high tumor mutation burden, and intratumoral lymphocytic infiltration. Programmed death-ligand 1 (PD-L1)/PD-1 signaling was described in solid tumor cells. In melanoma, liver, and thyroid cancer cells, intrinsic PD-1 signaling activates oncogenic functions, while in lung cancer cells, it has a tumor suppressor effect. Our work aimed to evaluate the effects of the anti-PD-1 nivolumab (NIVO) on CRC cells.
Methods: In vitro NIVO-treated human colon cancer cells (HT29, HCT116, and LoVo) were evaluated for cell growth, chemo/radiotherapeutic sensitivity, apoptosis, and spheroid growth. Total RNA-seq was assessed in 6-24 hours NIVO-treated human colon cancer cells HT29 and HCT116 as compared with NIVO-treated PES43 human melanoma cells. In vivo mice carrying HT29 xenograft were intraperitoneally treated with NIVO, OXA (oxaliplatin), and NIVO+OXA, and the tumors were characterized for growth, apoptosis, and pERK1/2/pP38. Forty-eight human primary colon cancers were evaluated for PD-1 expression through immunohistochemistry.
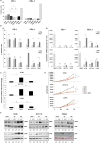
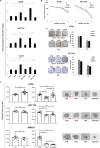
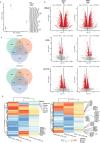
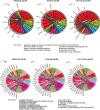
Results: In PD-1+ human colon cancer cells, intrinsic PD-1 signaling significantly decreased proliferation and promoted apoptosis. On the contrary, NIVO promoted proliferation, reduced apoptosis, and protected PD-1+ cells from chemo/radiotherapy. Transcriptional profile of NIVO-treated HT29 and HCT116 human colon cancer cells revealed downregulation of BATF2, DRAM1, FXYD3, IFIT3, MT-TN, and TNFRSF11A, and upregulation of CLK1, DCAF13, DNAJC2, MTHFD1L, PRPF3, PSMD7, and SCFD1; the opposite regulation was described in NIVO-treated human melanoma PES43 cells. Differentially expressed genes (DEGs) were significantly enriched for interferon pathway, innate immune, cytokine-mediated signaling pathways. In vivo, NIVO promoted HT29 tumor growth, thus reducing OXA efficacy as revealed through significant Ki-67 increase, pERK1/2 and pP38 increase, and apoptotic cell reduction. Eleven out of 48 primary human colon cancer biopsies expressed PD-1 (22.9%). PD-1 expression is significantly associated with lower pT stage.
Conclusions: In PD-1+ human colon cancer cells, NIVO activates tumor survival pathways and could protect tumor cells from conventional therapies.
Keywords: gastrointestinal neoplasms; programmed cell death 1 receptor.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
