Mechanical control of nuclear import by Importin-7 is regulated by its dominant cargo YAP
- PMID: 35246520
- PMCID: PMC8897400
- DOI: 10.1038/s41467-022-28693-y
Mechanical control of nuclear import by Importin-7 is regulated by its dominant cargo YAP
Abstract
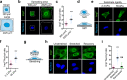
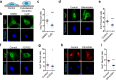
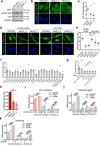
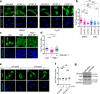
Mechanical forces regulate multiple essential pathways in the cell. The nuclear translocation of mechanoresponsive transcriptional regulators is an essential step for mechanotransduction. However, how mechanical forces regulate the nuclear import process is not understood. Here, we identify a highly mechanoresponsive nuclear transport receptor (NTR), Importin-7 (Imp7), that drives the nuclear import of YAP, a key regulator of mechanotransduction pathways. Unexpectedly, YAP governs the mechanoresponse of Imp7 by forming a YAP/Imp7 complex that responds to mechanical cues through the Hippo kinases MST1/2. Furthermore, YAP behaves as a dominant cargo of Imp7, restricting the Imp7 binding and the nuclear translocation of other Imp7 cargoes such as Smad3 and Erk2. Thus, the nuclear import process is an additional regulatory layer indirectly regulated by mechanical cues, which activate a preferential Imp7 cargo, YAP, which competes out other cargoes, resulting in signaling crosstalk.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1.EMBO J. 1999 May 4;18(9):2411-23. doi: 10.1093/emboj/18.9.2411. EMBO J. 1999. PMID: 10228156 Free PMC article.
-
Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication.J Biol Chem. 2007 May 4;282(18):13456-67. doi: 10.1074/jbc.M610546200. Epub 2007 Mar 14. J Biol Chem. 2007. PMID: 17360709
-
HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome.Retrovirology. 2009 Feb 4;6:11. doi: 10.1186/1742-4690-6-11. Retrovirology. 2009. PMID: 19193229 Free PMC article.
-
Nuclear import of histones.Biochem Soc Trans. 2020 Dec 18;48(6):2753-2767. doi: 10.1042/BST20200572. Biochem Soc Trans. 2020. PMID: 33300986 Free PMC article. Review.
-
Allosteric control of the exportin CRM1 unraveled by crystal structure analysis.FEBS J. 2014 Sep;281(18):4179-94. doi: 10.1111/febs.12842. Epub 2014 Jun 6. FEBS J. 2014. PMID: 24823279 Free PMC article. Review.
Cited by
-
NatB Protects Procaspase-8 from UBR4-Mediated Degradation and Is Required for Full Induction of the Extrinsic Apoptosis Pathway.Mol Cell Biol. 2024;44(9):358-371. doi: 10.1080/10985549.2024.2382453. Epub 2024 Aug 4. Mol Cell Biol. 2024. PMID: 39099191 Free PMC article.
-
Recognition motifs for importin 4 [(L)PPRS(G/P)P] and importin 5 [KP(K/Y)LV] binding, identified by bio-informatic simulation and experimental in vitro validation.Comput Struct Biotechnol J. 2022 Oct 26;20:5952-5961. doi: 10.1016/j.csbj.2022.10.015. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36382187 Free PMC article.
-
Nuclear Import and Export of YAP and TAZ.Cancers (Basel). 2023 Oct 12;15(20):4956. doi: 10.3390/cancers15204956. Cancers (Basel). 2023. PMID: 37894323 Free PMC article. Review.
-
Depletion of SUN1/2 induces heterochromatin accrual in mesenchymal stem cells during adipogenesis.Commun Biol. 2025 Mar 13;8(1):428. doi: 10.1038/s42003-025-07832-3. Commun Biol. 2025. PMID: 40082539 Free PMC article.
-
Modulation of Gene Expression by Substrate Stiffness via Ubiquitination of Histone H2B by Ubiquitin-Conjugating Enzyme E2A/B.ACS Omega. 2025 Apr 9;10(15):15799-15809. doi: 10.1021/acsomega.5c02459. eCollection 2025 Apr 22. ACS Omega. 2025. PMID: 40290990 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

