Cytonemes coordinate asymmetric signaling and organization in the Drosophila muscle progenitor niche
- PMID: 35246530
- PMCID: PMC8897416
- DOI: 10.1038/s41467-022-28587-z
Cytonemes coordinate asymmetric signaling and organization in the Drosophila muscle progenitor niche
Abstract
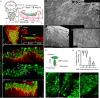
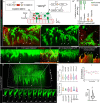
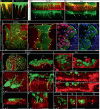
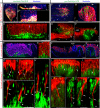
Asymmetric signaling and organization in the stem-cell niche determine stem-cell fates. Here, we investigate the basis of asymmetric signaling and stem-cell organization using the Drosophila wing-disc that creates an adult muscle progenitor (AMP) niche. We show that AMPs extend polarized cytonemes to contact the disc epithelial junctions and adhere themselves to the disc/niche. Niche-adhering cytonemes localize FGF-receptor to selectively adhere to the FGF-producing disc and receive FGFs in a contact-dependent manner. Activation of FGF signaling in AMPs, in turn, reinforces disc-specific cytoneme polarity/adhesion, which maintains their disc-proximal positions. Loss of cytoneme-mediated adhesion promotes AMPs to lose niche occupancy and FGF signaling, occupy a disc-distal position, and acquire morphological hallmarks of differentiation. Niche-specific AMP organization and diversification patterns are determined by localized expression and presentation patterns of two different FGFs in the wing-disc and their polarized target-specific distribution through niche-adhering cytonemes. Thus, cytonemes are essential for asymmetric signaling and niche-specific AMP organization.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

