Recombinant human interleukin-7 reverses T cell exhaustion ex vivo in critically ill COVID-19 patients
- PMID: 35246776
- PMCID: PMC8896969
- DOI: 10.1186/s13613-022-00982-1
Recombinant human interleukin-7 reverses T cell exhaustion ex vivo in critically ill COVID-19 patients
Erratum in
-
Correction to: Recombinant human interleukin-7 reverses T cell exhaustion ex vivo in critically ill COVID-19 patients.Ann Intensive Care. 2022 Apr 1;12(1):30. doi: 10.1186/s13613-022-01007-7. Ann Intensive Care. 2022. PMID: 35362873 Free PMC article. No abstract available.
Abstract
Background: Lymphopenia is a hallmark of severe coronavirus disease 19 (COVID-19). Similar alterations have been described in bacterial sepsis and therapeutic strategies targeting T cell function such as recombinant human interleukin 7 (rhIL-7) have been proposed in this clinical context. As COVID-19 is a viral sepsis, the objectives of this study were to characterize T lymphocyte response over time in severe COVID-19 patients and to assess the effect of ex vivo administration of rhIL-7.
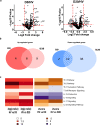
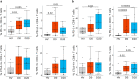
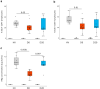
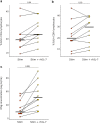
Results: Peripheral blood mononuclear cells from COVID-19 patients hospitalized in intensive care unit (ICU) were collected at admission and after 20 days. Transcriptomic profile was evaluated through NanoString technology. Inhibitory immune checkpoints expressions were determined by flow cytometry. T lymphocyte proliferation and IFN-γ production were evaluated after ex vivo stimulation in the presence or not of rhIL-7. COVID-19 ICU patients were markedly lymphopenic at admission. Mononuclear cells presented with inhibited transcriptomic profile prevalently with impaired T cell activation pathways. CD4 + and CD8 + T cells presented with over-expression of co-inhibitory molecules PD-1, PD-L1, CTLA-4 and TIM-3. CD4 + and CD8 + T cell proliferation and IFN-γ production were markedly altered in samples collected at ICU admission. These alterations, characteristic of a T cell exhaustion state, were more pronounced at ICU admission and alleviated over time. Treatment with rhIL-7 ex vivo significantly improved both T cell proliferation and IFN-γ production in cells from COVID-19 patients.
Conclusions: Severe COVID-19 patients present with features of profound T cell exhaustion upon ICU admission which can be reversed ex vivo by rhIL-7. These results reinforce our understanding of severe COVID-19 pathophysiology and opens novel therapeutic avenues to treat such critically ill patients based of immunomodulation approaches. Defining the appropriate timing for initiating such immune-adjuvant therapy in clinical setting and the pertinent markers for a careful selection of patients are now warranted to confirm the ex vivo results described so far. Trial registration ClinicalTrials.gov identifier: NCT04392401 Registered 18 May 2020, http:// clinicaltrials.gov/ct2/show/NCT04392401.
Keywords: Critically ill; Exhaustion; Immunostimulation; Interleukin-7; SARS-CoV-2; T lymphocytes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests in relation to the work. MAC, KBP and MM are bioMérieux’s employees. This private company had no role in the study design, result analysis and decision to publish this study.
Figures
References
-
- Carenzo L, Costantini E, Greco M, Barra FL, Rendiniello V, Mainetti M, et al. Hospital surge capacity in a tertiary emergency referral centre during the COVID-19 outbreak in Italy. Anaesthesia. 2020;75(7):928–934. - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

