Impact of structural dynamics on biological functions of flaviviruses
- PMID: 35246954
- PMCID: PMC10952610
- DOI: 10.1111/febs.16419
Impact of structural dynamics on biological functions of flaviviruses
Abstract
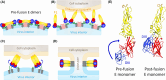
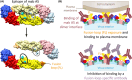
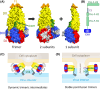
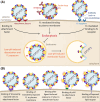
Flaviviruses comprise a number of mosquito- or tick-transmitted human pathogens of global public health importance. Advances in structural biology techniques have contributed substantially to our current understanding of the life cycle of these small enveloped RNA viruses and led to deep insights into details of virus assembly, maturation and cell entry. In addition to large-scale conformational changes and oligomeric rearrangements of envelope proteins during these processes, there is increasing evidence that smaller-scale protein dynamics (referred to as virus "breathing") can confer extra flexibility to these viruses for the fine-tuning of their interactions with the immune system and possibly with cellular factors they encounter in their complex ecological cycles in arthropod and vertebrate hosts. In this review, we discuss how work with tick-borne encephalitis virus has extended our view on flavivirus breathing, leading to the identification of a novel mechanism of antibody-mediated infection enhancement and demonstrating breathing intermediates of the envelope protein in the process of membrane fusion. These data are discussed in the context of other flaviviruses and the perspective of a potential role of virus breathing to cope with the requirements of adaptation and replication in evolutionarily very different hosts.
Keywords: antibody-dependent enhancement; envelope protein; flavivirus; membrane fusion; particle dynamics; particle heterogeneity; tick-borne encephalitis virus; virus breathing.
© 2022 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
None to declare with this review. The Center for Virology received a research grant from Pfizer on the epidemiology of TBE in Austria, with KS as principal investigator (2018‐2021). FXH and KS are inventors on a patent of the Medical University of Vienna on flavivirus IgM serodiagnosis.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

