Age-Related Changes in the Nasopharyngeal Microbiome Are Associated With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection and Symptoms Among Children, Adolescents, and Young Adults
- PMID: 35247047
- PMCID: PMC8903463
- DOI: 10.1093/cid/ciac184
Age-Related Changes in the Nasopharyngeal Microbiome Are Associated With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection and Symptoms Among Children, Adolescents, and Young Adults
Abstract
Background: Children are less susceptible to SARS-CoV-2 infection and typically have milder illness courses than adults, but the factors underlying these age-associated differences are not well understood. The upper respiratory microbiome undergoes substantial shifts during childhood and is increasingly recognized to influence host defense against respiratory pathogens. Thus, we sought to identify upper respiratory microbiome features associated with SARS-CoV-2 infection susceptibility and illness severity.
Methods: We collected clinical data and nasopharyngeal swabs from 285 children, adolescents, and young adults (<21 years) with documented SARS-CoV-2 exposure. We used 16S ribosomal RNA gene sequencing to characterize the nasopharyngeal microbiome and evaluated for age-adjusted associations between microbiome characteristics and SARS-CoV-2 infection status and respiratory symptoms.
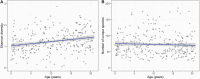
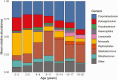
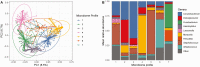
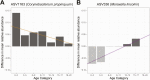
Results: Nasopharyngeal microbiome composition varied with age (PERMANOVA, P < .001; R2 = 0.06) and between SARS-CoV-2-infected individuals with and without respiratory symptoms (PERMANOVA, P = .002; R2 = 0.009). SARS-CoV-2-infected participants with Corynebacterium/Dolosigranulum-dominant microbiome profiles were less likely to have respiratory symptoms than infected participants with other nasopharyngeal microbiome profiles (OR: .38; 95% CI: .18-.81). Using generalized joint attributed modeling, we identified 9 bacterial taxa associated with SARS-CoV-2 infection and 6 taxa differentially abundant among SARS-CoV-2-infected participants with respiratory symptoms; the magnitude of these associations was strongly influenced by age.
Conclusions: We identified interactive relationships between age and specific nasopharyngeal microbiome features that are associated with SARS-CoV-2 infection susceptibility and symptoms in children, adolescents, and young adults. Our data suggest that the upper respiratory microbiome may be a mechanism by which age influences SARS-CoV-2 susceptibility and illness severity.
Keywords: Corynebacterium; Dolosigranulum; COVID-19; generalized joint attribute modeling; pediatric microbiota.
© The Author(s) 2022. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures
Update of
-
Age-related changes in the upper respiratory microbiome are associated with SARS-CoV-2 susceptibility and illness severity.medRxiv [Preprint]. 2021 Mar 23:2021.03.20.21252680. doi: 10.1101/2021.03.20.21252680. medRxiv. 2021. Update in: Clin Infect Dis. 2022 Aug 24;75(1):e928-e937. doi: 10.1093/cid/ciac184. PMID: 33791716 Free PMC article. Updated. Preprint.
Similar articles
-
Age-related changes in the upper respiratory microbiome are associated with SARS-CoV-2 susceptibility and illness severity.medRxiv [Preprint]. 2021 Mar 23:2021.03.20.21252680. doi: 10.1101/2021.03.20.21252680. medRxiv. 2021. Update in: Clin Infect Dis. 2022 Aug 24;75(1):e928-e937. doi: 10.1093/cid/ciac184. PMID: 33791716 Free PMC article. Updated. Preprint.
-
SARS-CoV-2 infection and viral load are associated with the upper respiratory tract microbiome.J Allergy Clin Immunol. 2021 Apr;147(4):1226-1233.e2. doi: 10.1016/j.jaci.2021.02.001. Epub 2021 Feb 9. J Allergy Clin Immunol. 2021. PMID: 33577896 Free PMC article.
-
Longitudinal dynamics of the nasopharyngeal microbiome in response to SARS-CoV-2 Omicron variant and HIV infection in Kenyan women and their children.mSystems. 2025 May 20;10(5):e0156824. doi: 10.1128/msystems.01568-24. Epub 2025 Apr 22. mSystems. 2025. PMID: 40261064 Free PMC article.
-
The role of respiratory microbiota in the protection against viral diseases: respiratory commensal bacteria as next-generation probiotics for COVID-19.Biosci Microbiota Food Health. 2022;41(3):94-102. doi: 10.12938/bmfh.2022-009. Epub 2022 Mar 29. Biosci Microbiota Food Health. 2022. PMID: 35846832 Free PMC article. Review.
-
Human and novel coronavirus infections in children: a review.Paediatr Int Child Health. 2021 Feb;41(1):36-55. doi: 10.1080/20469047.2020.1781356. Epub 2020 Jun 25. Paediatr Int Child Health. 2021. PMID: 32584199 Review.
Cited by
-
Airway and Oral microbiome profiling of SARS-CoV-2 infected asthma and non-asthma cases revealing alterations-A pulmonary microbial investigation.PLoS One. 2023 Aug 17;18(8):e0289891. doi: 10.1371/journal.pone.0289891. eCollection 2023. PLoS One. 2023. PMID: 37590197 Free PMC article.
-
Characterization of upper airway microbiome across severity of COVID-19 during hospitalization and treatment.Front Cell Infect Microbiol. 2023 Jul 4;13:1205401. doi: 10.3389/fcimb.2023.1205401. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37469595 Free PMC article.
-
Microbiome in the nasopharynx: Insights into the impact of COVID-19 severity.Heliyon. 2024 May 18;10(10):e31562. doi: 10.1016/j.heliyon.2024.e31562. eCollection 2024 May 30. Heliyon. 2024. PMID: 38826746 Free PMC article.
-
Guidance for prevention and management of COVID-19 in children and adolescents: A consensus statement from the Pediatric Infectious Diseases Society Pediatric COVID-19 Therapies Taskforce.J Pediatric Infect Dis Soc. 2024 Mar 19;13(3):159-185. doi: 10.1093/jpids/piad116. J Pediatric Infect Dis Soc. 2024. PMID: 38339996 Free PMC article.
-
Impact of age on pneumococcal colonization of the nasopharynx and oral cavity: an ecological perspective.ISME Commun. 2024 Jan 12;4(1):ycae002. doi: 10.1093/ismeco/ycae002. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38390521 Free PMC article.
References
-
- Castagnoli R, Votto M, Licari A, et al. . Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children and adolescents: a systematic review. JAMA Pediatr 2020; 174:882–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

