Early reduction of SARS-CoV-2-replication in bronchial epithelium by kinin B2 receptor antagonism
- PMID: 35247068
- PMCID: PMC8897552
- DOI: 10.1007/s00109-022-02182-7
Early reduction of SARS-CoV-2-replication in bronchial epithelium by kinin B2 receptor antagonism
Abstract
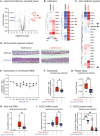
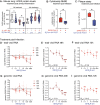
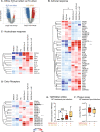
SARS-CoV-2 has evolved to enter the host via the ACE2 receptor which is part of the kinin-kallikrein pathway. This complex pathway is only poorly understood in context of immune regulation but critical to control infection. This study examines SARS-CoV-2-infection and epithelial mechanisms of the kinin-kallikrein-system at the kinin B2 receptor level in SARS-CoV-2-infection that is of direct translational relevance. From acute SARS-CoV-2-positive study participants and -negative controls, transcriptomes of nasal curettages were analyzed. Primary airway epithelial cells (NHBEs) were infected with SARS-CoV-2 and treated with the approved B2R-antagonist icatibant. SARS-CoV-2 RNA RT-qPCR, cytotoxicity assays, plaque assays, and transcriptome analyses were performed. The treatment effect was further studied in a murine airway inflammation model in vivo. Here, we report a broad and strong upregulation of kallikreins and the kinin B2 receptor (B2R) in the nasal mucosa of acutely symptomatic SARS-CoV-2-positive study participants. A B2R-antagonist impeded SARS-CoV-2 replication and spread in NHBEs, as determined in plaque assays on Vero-E6 cells. B2R-antagonism reduced the expression of SARS-CoV-2 entry receptor ACE2, G protein-coupled receptor signaling, and ion transport in vitro and in a murine airway inflammation in vivo model. In summary, this study provides evidence that treatment with B2R-antagonists protects airway epithelial cells from SARS-CoV-2 by inhibiting its replication and spread, through the reduction of ACE2 levels and the interference with several cellular signaling processes. Future clinical studies need to shed light on the airway protection potential of approved B2R-antagonists, like icatibant, in the treatment of early-stage COVID-19. KEY MESSAGES: Induction of kinin B2 receptor in the nose of SARS-CoV-2-positive patients. Treatment with B2R-antagonist protects airway epithelial cells from SARS-CoV-2. B2R-antagonist reduces ACE2 levels in vivo and ex vivo. Protection by B2R-antagonist is mediated by inhibiting viral replication and spread.
Keywords: ACE2; B2R-antagonist; COVID-19; Kinin; Kinin-kallikrein-system; SARS-CoV-2.
© 2022. The Author(s).
Conflict of interest statement
Dr. Jakwerth reports grants from Federal Ministry of Education and Research, grants from European Institute of Innovation & Technology (EIT), during the conduct of the study. Mr. Feuerherd reports personal fees from Helmholtz Zentrum München and Dr. Höhnle AG, outside the submitted work. Dr. Spinner reports grants, personal fees, non-financial support, and other from AbbVie, grants, personal fees, non-financial support, and other from Apeiron, personal fees from Formycon, grants, personal fees, non-financial support, and other from Gilead Sciences, grants, personal fees, and other from Eli Lilly, grants, personal fees, non-financial support, and other from Janssen-Cilag, grants, personal fees, non-financial support, and other from GSK/ViiV Healthcare, grants, personal fees, non-financial support, and other from MSD, outside the submitted work. Prof. Dr. Buch reports personal fees from Virometix AG, other from Virometix AG, other from Trials24 GmbH, other from Clemedi AG, outside the submitted work. Prof. Dr. Protzer reports grants from the Federal Ministry of Education and Research, the German Center for Infection Research (DZIF), the German Research Foundation (DFG), the European Union via Horizon 2020, and the Bavarian Research Foundation during conduct of the study. She receives personal fees as an ad hoc scientific advisor from Abbvie, Arbutus, Gilead, GSK, Johnson & Johnson, Vaccitech. Prof. Dr. Schmidt-Weber reports grants from German Center for Lung Research (DZL), grants from Comprehensive Pulmonary Lung Center (CPC) Munich during the conduct of the study. Personal fees from Allergopharma and Bencard, outside the submitted work. Dr. Zissler reports grants from Federal Ministry of Education and Research, during the conduct of the study. Dr. Chaker reports grants for clinical studies and research and other from Allergopharma, ALK Abello, AstraZeneca, Bencard / Allergen Therapeutics, ASIT Biotech, Lofarma, GSK, Novartis, LETI, Inmunotek, Roche, Sanofi Genzyme, Zeller, and from the European Institute of Technology (EIT); has received travel support from the European Academy of Allergy and Clinical Immunology (EAACI), DGAKI, all outside the submitted work. In addition, Drs. Jakwerth, Feuerherd, Protzer, Schmidt-Weber, Zissler, and Chaker are named as inventors on the patent application “Novel approaches for treatment of SARS-CoV-2-Infection in a patient.” Mr. Guerth, Ms. Oelsner, Dr. Schellhammer, Ms. Giglberger, Ms. Pechtold, Dr. Jerin, Ms. Kugler, Dr. Mogler, Dr. Haller, Ms. Erb, and Prof. Dr. Wollenberg have nothing to disclose.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

