Quantitative redox proteomics revealed molecular mechanisms of salt tolerance in the roots of sugar beet monomeric addition line M14
- PMID: 35247135
- PMCID: PMC8898211
- DOI: 10.1186/s40529-022-00337-w
Quantitative redox proteomics revealed molecular mechanisms of salt tolerance in the roots of sugar beet monomeric addition line M14
Abstract
Background: Salt stress is often associated with excessive production of reactive oxygen species (ROS). Oxidative stress caused by the accumulation of ROS is a major factor that negatively affects crop growth and yield. Root is the primary organ that senses and transmits the salt stress signal to the whole plant. How oxidative stress affect redox sensitive proteins in the roots is not known.
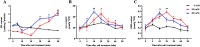
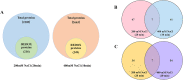
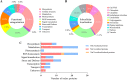
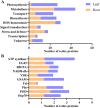
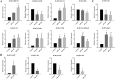
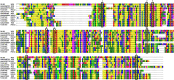
Results: In this study, the redox proteome of sugar beet M14 roots under salt stress was investigated. Using iTRAQ reporters, we determined that salt stress caused significant changes in the abundance of many proteins (2305 at 20 min salt stress and 2663 at 10 min salt stress). Using iodoTMT reporters, a total of 95 redox proteins were determined to be responsive to salt stress after normalizing again total protein level changes. Notably, most of the differential redox proteins were involved in metabolism, ROS homeostasis, and stress and defense, while a small number play a role in transport, biosynthesis, signal transduction, transcription and photosynthesis. Transcription levels of 14 genes encoding the identified redox proteins were analyzed using qRT-PCR. All the genes were induced by salt stress at the transcriptional level.
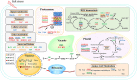
Conclusions: Based on the redox proteomics results, we construct a map of the regulatory network of M14 root redox proteins in response to salt stress. This study further refines the molecular mechanism of salt resistance at the level of protein redox regulation.
Keywords: Molecular mechanisms; Redox proteomics; Salt stress; Sugar beet M14; iodoTMTRAQ.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Cys-SH based quantitative redox proteomics of salt induced response in sugar beet monosomic addition line M14.Bot Stud. 2021 Oct 18;62(1):16. doi: 10.1186/s40529-021-00320-x. Bot Stud. 2021. PMID: 34661775 Free PMC article.
-
Phosphoproteomics revealed salt stress tolerance mechanisms in the roots of sugar beet monomeric addition line M14.J Proteomics. 2025 Sep 15;320:105488. doi: 10.1016/j.jprot.2025.105488. Epub 2025 Jul 12. J Proteomics. 2025. PMID: 40659243
-
Quantitative proteomics and phosphoproteomics of sugar beet monosomic addition line M14 in response to salt stress.J Proteomics. 2016 Jun 30;143:286-297. doi: 10.1016/j.jprot.2016.04.011. Epub 2016 May 24. J Proteomics. 2016. PMID: 27233743
-
Salt stress response of membrane proteome of sugar beet monosomic addition line M14.J Proteomics. 2015 Sep 8;127(Pt A):18-33. doi: 10.1016/j.jprot.2015.03.025. Epub 2015 Apr 3. J Proteomics. 2015. PMID: 25845583 Review.
-
Advances in Understanding the Physiological and Molecular Responses of Sugar Beet to Salt Stress.Front Plant Sci. 2019 Nov 6;10:1431. doi: 10.3389/fpls.2019.01431. eCollection 2019. Front Plant Sci. 2019. PMID: 31781145 Free PMC article. Review.
Cited by
-
Salt Tolerance in Sugar Beet: From Impact Analysis to Adaptive Mechanisms and Future Research.Plants (Basel). 2024 Oct 28;13(21):3018. doi: 10.3390/plants13213018. Plants (Basel). 2024. PMID: 39519937 Free PMC article. Review.
-
Comparative proteomic analysis on chloroplast proteins provides new insights into the effects of low temperature in sugar beet.Bot Stud. 2022 Jun 7;63(1):18. doi: 10.1186/s40529-022-00349-6. Bot Stud. 2022. PMID: 35670889 Free PMC article.
-
Comparative Ubiquitination Proteomics Revealed the Salt Tolerance Mechanism in Sugar Beet Monomeric Additional Line M14.Int J Mol Sci. 2022 Dec 17;23(24):16088. doi: 10.3390/ijms232416088. Int J Mol Sci. 2022. PMID: 36555729 Free PMC article.
-
Effects of Salt Stress on the Morphology, Growth and Physiological Parameters of Juglansmicrocarpa L. Seedlings.Plants (Basel). 2022 Sep 12;11(18):2381. doi: 10.3390/plants11182381. Plants (Basel). 2022. PMID: 36145780 Free PMC article.
-
Integrative Multi-PTM Proteomics Reveals Dynamic Global, Redox, Phosphorylation, and Acetylation Regulation in Cytokine-Treated Pancreatic Beta Cells.Mol Cell Proteomics. 2024 Dec;23(12):100881. doi: 10.1016/j.mcpro.2024.100881. Epub 2024 Nov 15. Mol Cell Proteomics. 2024. PMID: 39550035 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

