The evolutionarily conserved arginyltransferase 1 mediates a pVHL-independent oxygen-sensing pathway in mammalian cells
- PMID: 35247316
- PMCID: PMC8957288
- DOI: 10.1016/j.devcel.2022.02.010
The evolutionarily conserved arginyltransferase 1 mediates a pVHL-independent oxygen-sensing pathway in mammalian cells
Abstract
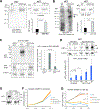
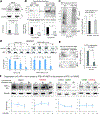
The response to oxygen availability is a fundamental process concerning metabolism and survival/death in all mitochondria-containing eukaryotes. However, the known oxygen-sensing mechanism in mammalian cells depends on pVHL, which is only found among metazoans but not in other species. Here, we present an alternative oxygen-sensing pathway regulated by ATE1, an enzyme ubiquitously conserved in eukaryotes that influences protein degradation by posttranslational arginylation. We report that ATE1 centrally controls the hypoxic response and glycolysis in mammalian cells by preferentially arginylating HIF1α that is hydroxylated by PHD in the presence of oxygen. Furthermore, the degradation of arginylated HIF1α is independent of pVHL E3 ubiquitin ligase but dependent on the UBR family proteins. Bioinformatic analysis of human tumor data reveals that the ATE1/UBR and pVHL pathways jointly regulate oxygen sensing in a transcription-independent manner with different tissue specificities. Phylogenetic analysis suggests that eukaryotic ATE1 likely evolved during mitochondrial domestication, much earlier than pVHL.
Keywords: ATE1; Arginylation; HIF1α; Warburg effect; arginyltransferase; degradation; glycolysis; hypoxic signaling; oxygen sensing; posttranslational protein modification.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflict of interest.
Figures





Similar articles
-
Analyzing N-terminal Arginylation through the Use of Peptide Arrays and Degradation Assays.J Biol Chem. 2016 Sep 30;291(40):20976-20992. doi: 10.1074/jbc.M116.747956. Epub 2016 Aug 10. J Biol Chem. 2016. PMID: 27510035 Free PMC article.
-
Arginyltransferase 1 (ATE1)-mediated proteasomal degradation of viral haemagglutinin protein: a unique host defence mechanism.J Gen Virol. 2024 Aug;105(8). doi: 10.1099/jgv.0.002020. J Gen Virol. 2024. PMID: 39207120
-
Posttranslational arginylation enzyme Ate1 affects DNA mutagenesis by regulating stress response.Cell Death Dis. 2016 Sep 29;7(9):e2378. doi: 10.1038/cddis.2016.284. Cell Death Dis. 2016. PMID: 27685622 Free PMC article.
-
Post-translational protein arginylation in the normal nervous system and in neurodegeneration.J Neurochem. 2016 Aug;138(4):506-17. doi: 10.1111/jnc.13708. Epub 2016 Jul 5. J Neurochem. 2016. PMID: 27318192 Review.
-
Protein arginylation, a global biological regulator that targets actin cytoskeleton and the muscle.Anat Rec (Hoboken). 2014 Sep;297(9):1630-6. doi: 10.1002/ar.22969. Anat Rec (Hoboken). 2014. PMID: 25125176 Free PMC article. Review.
Cited by
-
The lower expression of circulating miR-210 and elevated serum levels of HIF-1α in ischemic stroke; Possible markers for diagnosis and disease prediction.J Clin Lab Anal. 2021 Dec;35(12):e24073. doi: 10.1002/jcla.24073. Epub 2021 Oct 28. J Clin Lab Anal. 2021. PMID: 34708885 Free PMC article.
-
Arginyltransferase1 drives a mitochondria-dependent program to induce cell death.bioRxiv [Preprint]. 2024 Nov 23:2024.11.22.624728. doi: 10.1101/2024.11.22.624728. bioRxiv. 2024. Update in: Cell Death Dis. 2025 Aug 16;16(1):622. doi: 10.1038/s41419-025-07917-1. PMID: 39605427 Free PMC article. Updated. Preprint.
-
Assaying Arginylation Activity in Cell Lysates Using a Fluorescent Reporter.Methods Mol Biol. 2023;2620:71-80. doi: 10.1007/978-1-0716-2942-0_9. Methods Mol Biol. 2023. PMID: 37010750
-
The N-degron pathway mediates lipophagy: The chemical modulation of lipophagy in obesity and NAFLD.Metabolism. 2023 Sep;146:155644. doi: 10.1016/j.metabol.2023.155644. Epub 2023 Jun 28. Metabolism. 2023. PMID: 37385404 Free PMC article.
-
Editorial: Waken the Silent Majority: Principles and Pathogenic Significance of Non-Acetyl Acylation and Other Understudied Post-Translational Modifications.Front Cell Dev Biol. 2022 Apr 13;10:896324. doi: 10.3389/fcell.2022.896324. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35493078 Free PMC article. No abstract available.
References
-
- ANGLARD P, TRAHAN E, LIU S, LATIF F, MERINO MJ, LERMAN MI, ZBAR B & LINEHAN WM 1992. Molecular and Cellular Characterization of Human Renal-Cell Carcinoma Cell-Lines. Cancer Res, 52, 348–356. - PubMed
-
- BACHMAIR A, FINLEY D & VARSHAVSKY A 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science, 234, 179–86. - PubMed
-
- BALZI E, CHODER M, CHEN WN, VARSHAVSKY A & GOFFEAU A 1990. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem, 265, 7464–71. - PubMed

