Tumor Intrinsic PD-L1 Promotes DNA Repair in Distinct Cancers and Suppresses PARP Inhibitor-Induced Synthetic Lethality
- PMID: 35247877
- PMCID: PMC9987177
- DOI: 10.1158/0008-5472.CAN-21-2076
Tumor Intrinsic PD-L1 Promotes DNA Repair in Distinct Cancers and Suppresses PARP Inhibitor-Induced Synthetic Lethality
Abstract
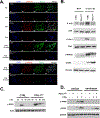
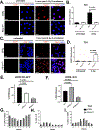
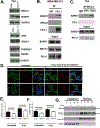
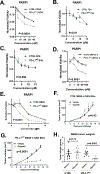
BRCA1-mediated homologous recombination is an important DNA repair mechanism that is the target of FDA-approved PARP inhibitors, yet details of BRCA1-mediated functions remain to be fully elucidated. Similarly, immune checkpoint molecules are targets of FDA-approved cancer immunotherapies, but the biological and mechanistic consequences of their application are incompletely understood. We show here that the immune checkpoint molecule PD-L1 regulates homologous recombination in cancer cells by promoting BRCA1 nuclear foci formation and DNA end resection. Genetic depletion of tumor PD-L1 reduced homologous recombination, increased nonhomologous end joining, and elicited synthetic lethality to PARP inhibitors olaparib and talazoparib in vitro in some, but not all, BRCA1 wild-type tumor cells. In vivo, genetic depletion of tumor PD-L1 rendered olaparib-resistant tumors sensitive to olaparib. In contrast, anti-PD-L1 immune checkpoint blockade neither enhanced olaparib synthetic lethality nor improved its efficacy in vitro or in wild-type mice. Tumor PD-L1 did not alter expression of BRCA1 or its cofactor BARD1 but instead coimmunoprecipitated with BARD1 and increased BRCA1 nuclear accumulation. Tumor PD-L1 depletion enhanced tumor CCL5 expression and TANK-binding kinase 1 activation in vitro, similar to known immune-potentiating effects of PARP inhibitors. Collectively, these data define immune-dependent and immune-independent effects of PARP inhibitor treatment and genetic tumor PD-L1 depletion. Moreover, they implicate a tumor cell-intrinsic, immune checkpoint-independent function of PD-L1 in cancer cell BRCA1-mediated DNA damage repair with translational potential, including as a treatment response biomarker.
Significance: PD-L1 upregulates BRCA1-mediated homologous recombination, and PD-L1-deficient tumors exhibit BRCAness by manifesting synthetic lethality in response to PARP inhibitors, revealing an exploitable therapeutic vulnerability and a candidate treatment response biomarker. See related commentary by Hanks, p. 2069.
©2022 American Association for Cancer Research.
Conflict of interest statement
Figures

Comment in
-
The "Inside" Story on Tumor-Expressed PD-L1.Cancer Res. 2022 Jun 6;82(11):2069-2071. doi: 10.1158/0008-5472.CAN-22-1060. Cancer Res. 2022. PMID: 35661198 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

