Single-cell Characterization of the Cellular Landscape of Acral Melanoma Identifies Novel Targets for Immunotherapy
- PMID: 35247927
- PMCID: PMC9106889
- DOI: 10.1158/1078-0432.CCR-21-3145
Single-cell Characterization of the Cellular Landscape of Acral Melanoma Identifies Novel Targets for Immunotherapy
Abstract
Purpose: Acral melanoma is a rare subtype of melanoma that arises on the non-hair-bearing skin of the palms, soles, and nail beds. In this study, we used single-cell RNA sequencing (scRNA-seq) to map the transcriptional landscape of acral melanoma and identify novel immunotherapeutic targets.
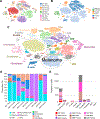
Experimental design: We performed scRNA-seq on nine clinical specimens (five primary, four metastases) of acral melanoma. Detailed cell type curation was performed, the immune landscapes were mapped, and key results were validated by analysis of The Cancer Genome Atlas (TCGA) and single-cell datasets. Cell-cell interactions were inferred and compared with those in nonacral cutaneous melanoma.
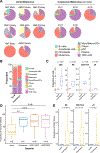
Results: Multiple phenotypic subsets of T cells, natural killer (NK) cells, B cells, macrophages, and dendritic cells with varying levels of activation/exhaustion were identified. A comparison between primary and metastatic acral melanoma identified gene signatures associated with changes in immune responses and metabolism. Acral melanoma was characterized by a lower overall immune infiltrate, fewer effector CD8 T cells and NK cells, and a near-complete absence of γδ T cells compared with nonacral cutaneous melanomas. Immune cells associated with acral melanoma exhibited expression of multiple checkpoints including PD-1, LAG-3, CTLA-4, V-domain immunoglobin suppressor of T cell activation (VISTA), TIGIT, and the Adenosine A2A receptor (ADORA2). VISTA was expressed in 58.3% of myeloid cells and TIGIT was expressed in 22.3% of T/NK cells.
Conclusions: Acral melanoma has a suppressed immune environment compared with that of cutaneous melanoma from nonacral skin. Expression of multiple, therapeutically tractable immune checkpoints were observed, offering new options for clinical translation.
©2022 American Association for Cancer Research.
Conflict of interest statement
Conflicts of interest:
Dr. Sarnaik is a co-inventor on a patent application with Provectus Biopharmaceuticals and has received consulting fees from Iovance Biotherapeutics, Guidepoint, Defined Health, Huron Consulting Group, KeyQuest Health Inc, and Gerson Lehrman Group and speaker fees from Physicians’ Educational Resource (PER) LLC, Medscape and Medstar Health. Dr. Sondak is a consultant for Bristol Myers Squibb, Eisai, Merck, Novartis, Regeneron and Replimune and receives research funding unrelated to this topic from Neogene Therapeutics. Dr. Koomen receives salary support from Bristol Myers Squibb unrelated to this work. Dr. Tarhini reports Consulting/Advisory Board Participation: Merck, Bristol Myers Squibb, Novartis, Genentech-Roche, Partner Therapeutics, Sanofi-Genzyme, Regeneron, Eisai and Clinigen. He also has support for contracted research from OncoSec, Clinigen, Genentech-Roche, Bristol Myers Squibb, Nektar Therapeutics, Sanofi-Genzyme, Regeneron, Navigate. Dr Khushalani is a paid advisor for Array Biopharma, Bristol Myers Squibb, EMD Serono, Genentech, HUYA Bioscience International, Nektar, Regeneron, Immunocore, Merck, Astra Zeneca, Incyte and Jounce. He receives research funding from HUYA Bioscience International, Regeneron, Merck, Amgen, Celgene, GlaxoSmithKline, Novartis, Replimune, and BMS. All other authors have no conflicts of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

