Activation of cryptic xylose metabolism by a transcriptional activator Znf1 boosts up xylitol production in the engineered Saccharomyces cerevisiae lacking xylose suppressor BUD21 gene
- PMID: 35248023
- PMCID: PMC8897867
- DOI: 10.1186/s12934-022-01757-w
Activation of cryptic xylose metabolism by a transcriptional activator Znf1 boosts up xylitol production in the engineered Saccharomyces cerevisiae lacking xylose suppressor BUD21 gene
Abstract
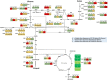
Background: Xylitol is a valuable pentose sugar alcohol, used in the food and pharmaceutical industries. Biotechnological xylitol production is currently attractive due to possible conversion from abundant and low-cost industrial wastes or agricultural lignocellulosic biomass. In this study, the transcription factor Znf1 was characterised as being responsible for the activation of cryptic xylose metabolism in a poor xylose-assimilating S. cerevisiae for xylitol production.
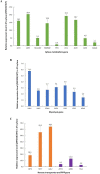
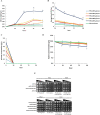
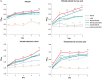
Results: The results suggest that the expression of several xylose-utilising enzyme genes, encoding xylose reductases for the reduction of xylose to xylitol was derepressed by xylose. Their expression and those of a pentose phosphate shunt and related pathways required for xylose utilisation were strongly activated by the transcription factor Znf1. Using an engineered S. cerevisiae strain overexpressing ZNF1 in the absence of the xylose suppressor bud21Δ, xylitol production was maximally by approximately 1200% to 12.14 g/L of xylitol, corresponding to 0.23 g/g xylose consumed, during 10% (w/v) xylose fermentation. Proteomic analysis supported the role of Znf1 and Bud21 in modulating levels of proteins associated with carbon metabolism, xylose utilisation, ribosomal protein synthesis, and others. Increased tolerance to lignocellulosic inhibitors and improved cell dry weight were also observed in this engineered bud21∆ + pLJ529-ZNF1 strain. A similar xylitol yield was achieved using fungus-pretreated rice straw hydrolysate as an eco-friendly and low-cost substrate.
Conclusions: Thus, we identified the key modulators of pentose sugar metabolism, namely the transcription factor Znf1 and the suppressor Bud21, for enhanced xylose utilisation, providing a potential application of a generally recognised as safe yeast in supporting the sugar industry and the sustainable lignocellulose-based bioeconomy.
Keywords: Bioconversion; Metabolic engineering; Transcription factor Znf1; Xylitol; Xylose utilisation; Yeast.
© 2022. The Author(s).
Figures




Similar articles
-
Increased production of isobutanol from xylose through metabolic engineering of Saccharomyces cerevisiae overexpressing transcription factor Znf1 and exogenous genes.FEMS Yeast Res. 2024 Jan 9;24:foae006. doi: 10.1093/femsyr/foae006. FEMS Yeast Res. 2024. PMID: 38331422 Free PMC article.
-
The impact of transcription factors Znf1, Sip4, Adr1, Tup1, and Hap4 on xylose alcoholic fermentation in the engineered yeast Saccharomyces cerevisiae.Antonie Van Leeuwenhoek. 2021 Sep;114(9):1373-1385. doi: 10.1007/s10482-021-01607-6. Epub 2021 Jun 25. Antonie Van Leeuwenhoek. 2021. PMID: 34170419
-
Reprogramming of the Ethanol Stress Response in Saccharomyces cerevisiae by the Transcription Factor Znf1 and Its Effect on the Biosynthesis of Glycerol and Ethanol.Appl Environ Microbiol. 2021 Jul 27;87(16):e0058821. doi: 10.1128/AEM.00588-21. Epub 2021 Jul 27. Appl Environ Microbiol. 2021. PMID: 34105981 Free PMC article.
-
Relation of xylitol formation and lignocellulose degradation in yeast.Appl Microbiol Biotechnol. 2023 May;107(10):3143-3151. doi: 10.1007/s00253-023-12495-3. Epub 2023 Apr 11. Appl Microbiol Biotechnol. 2023. PMID: 37039848 Review.
-
Biosynthetic strategies to produce xylitol: an economical venture.Appl Microbiol Biotechnol. 2019 Jul;103(13):5143-5160. doi: 10.1007/s00253-019-09881-1. Epub 2019 May 17. Appl Microbiol Biotechnol. 2019. PMID: 31101942 Review.
Cited by
-
Increased production of isobutanol from xylose through metabolic engineering of Saccharomyces cerevisiae overexpressing transcription factor Znf1 and exogenous genes.FEMS Yeast Res. 2024 Jan 9;24:foae006. doi: 10.1093/femsyr/foae006. FEMS Yeast Res. 2024. PMID: 38331422 Free PMC article.
-
Integrated omic analysis of a new flavor yeast strain in fermented rice milk.FEMS Yeast Res. 2025 Jan 30;25:foaf017. doi: 10.1093/femsyr/foaf017. FEMS Yeast Res. 2025. PMID: 40153366 Free PMC article.
-
Advances in the biological production of sugar alcohols from biomass-derived xylose.World J Microbiol Biotechnol. 2025 Mar 28;41(4):110. doi: 10.1007/s11274-025-04316-8. World J Microbiol Biotechnol. 2025. PMID: 40148723 Review.
-
New biomarkers underlying acetic acid tolerance in the probiotic yeast Saccharomyces cerevisiae var. boulardii.Appl Microbiol Biotechnol. 2024 Jan 19;108(1):153. doi: 10.1007/s00253-023-12946-x. Appl Microbiol Biotechnol. 2024. PMID: 38240846 Free PMC article.
-
Biosynthesis of D-threitol from biomass-derived xylose by engineering Trichosporonoides oedocephalis.World J Microbiol Biotechnol. 2025 Jul 4;41(7):248. doi: 10.1007/s11274-025-04425-4. World J Microbiol Biotechnol. 2025. PMID: 40613979
References
-
- Werpy T, Holladay J, White J: Top value added chemicals from biomass: I. Results of screening for potential candidates from sugars and synthesis gas. Pacific Northwest National Laboratory (PNNL) and National Renewable Energy Laboratory (NREL). 2004.
-
- Albuquerque TL, da Silva IJ, de Macedo GR, Rocha MVP. Biotechnological production of xylitol from lignocellulosic wastes: A review. Process Biochem. 2014;49:1779–1789.
-
- Dasgupta Q, Chatterjee K, Madras G. Combinatorial approach to develop tailored biodegradable poly(xylitol dicarboxylate) polyesters. Biomacromol. 2014;15:4302–4313. - PubMed
-
- Shankar K, Kulkarni NS, Sajjanshetty R, Jayalakshmi SK, Sreeramulu K. Co-production of xylitol and ethanol by the fermentation of the lignocellulosic hydrolysates of banana and water hyacinth leaves by individual yeast strains. Ind Crops Prod. 2020;155:112809.
-
- Mohamad NL, Mustapa Kamal S, Mokhtar M. Xylitol biological production: A review of recent studies. Food Rev Int. 2015;31:74–89.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

