Detection and quantification of SARS-CoV-2 RNA in wastewater influent in relation to reported COVID-19 incidence in Finland
- PMID: 35248908
- PMCID: PMC8865022
- DOI: 10.1016/j.watres.2022.118220
Detection and quantification of SARS-CoV-2 RNA in wastewater influent in relation to reported COVID-19 incidence in Finland
Abstract
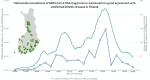
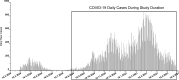
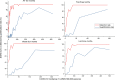
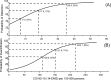
Wastewater-based surveillance is a cost-effective concept for monitoring COVID-19 pandemics at a population level. Here, SARS-CoV-2 RNA was monitored from a total of 693 wastewater (WW) influent samples from 28 wastewater treatment plants (WWTP, N = 21-42 samples per WWTP) in Finland from August 2020 to May 2021, covering WW of ca. 3.3 million inhabitants (∼ 60% of the Finnish population). Quantity of SARS-CoV-2 RNA fragments in 24 h-composite samples was determined by using the ultrafiltration method followed by nucleic acid extraction and CDC N2 RT-qPCR assay. SARS-CoV-2 RNA signals at each WWTP were compared over time to the numbers of confirmed COVID-19 cases (14-day case incidence rate) in the sewer network area. Over the 10-month surveillance period with an extensive total number of samples, the detection rate of SARS-CoV-2 RNA in WW was 79% (including 6% uncertain results, i.e., amplified only in one out of four, two original and two ten-fold diluted replicates), while only 24% of all samples exhibited gene copy numbers above the quantification limit. The range of the SARS-CoV-2 detection rate in WW varied from 33% (including 10% uncertain results) in Pietarsaari to 100% in Espoo. Only six out of 693 WW samples were positive with SARS-COV-2 RNA when the reported COVID-19 case number from the preceding 14 days was zero. Overall, the 14-day COVID-19 incidence was 7.0, 18, and 36 cases per 100 000 persons within the sewer network area when the probability to detect SARS-CoV-2 RNA in wastewater samples was 50%, 75% and 95%, respectively. The quantification of SARS-CoV-2 RNA required significantly more COVID-19 cases: the quantification rate was 50%, 75%, and 95% when the 14-day incidence was 110, 152, and 223 COVID-19 cases, respectively, per 100 000 persons. Multiple linear regression confirmed the relationship between the COVID-19 incidence and the SARS-CoV-2 RNA quantified in WW at 15 out of 28 WWTPs (overall R2 = 0.36, p < 0.001). At four of the 13 WWTPs where a significant relationship was not found, the SARS-CoV-2 RNA remained below the quantification limit during the whole study period. In the five other WWTPs, the sewer coverage was less than 80% of the total population in the area and thus the COVID-19 cases may have been inhabitants from the areas not covered. Based on the results obtained, WW-based surveillance of SARS-CoV-2 could be used as an indicator for local and national COVID-19 incidence trends. Importantly, the determination of SARS-CoV-2 RNA fragments from WW is a powerful and non-invasive public health surveillance measure, independent of possible changes in the clinical testing strategies or in the willingness of individuals to be tested for COVID-19.
Keywords: Community sewage; Coronavirus; National surveillance; RT-qPCR assay; Wastewater-based epidemiology.
Copyright © 2022. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020 doi: 10.1016/J.SCITOTENV.2020.138764. Doi. - DOI - PMC - PubMed
-
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O'brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. Doi. - DOI - PMC - PubMed
-
- Ahmed W., Simpson S., Bertsch P., Bibby K., Bivins A., Blackall L., Bofill-Mas S., Bosch A., Brandao J., Choi P., Ciesielski M., Donner E., D'Souza N., Farnleitner A., Gerrity D., Gonzalez R., Griffith J., Gyawali P., Haas C., Hamilton K., Hapuarachchi C., Harwood V., Haque R., Jackson G., Khan S., Khan W., Kitajima M., Korajkic A., La Rosa G., Layton B., Lipp E., McLellan S., McMinn B., Medema G., Metcalfe S., Meijer W., Mueller J., Murphy H., Naughton C., Noble R., Payyappat S., Petterson S., Pitkänen T., Rajal V., Reyneke B., Roman F., Rose J., Rusinol M., Sadowsky M., Sala-Comorera L., Setoh Y.X., Sherchan S., Sirikanchana K., Smith W., Steele J., Sabburg R., Symonds E., Thai P., Thomas K., Tynan J., Toze S., Thompson J., Whiteley A., Wong J., Sano D., Wuertz S., Xagoraraki I., Zhang Q., Zimmer-Faust A., Shanks O. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2021 doi: 10.1016/j.scitotenv.2021.149877. Doi. - DOI - PMC - PubMed
-
- Ahmed W., Bibby K., D'Aoust P.M., Delatolla R., Gerba C.P., Haas C.N., Hamilton K.A., Hewitt J., Julian T.R., Kaya D., Monis P., Moulin L., Naughton C., Noble R.T., Shrestha A., Tiwari A., Simpson S.L., Wurtzer S., Bivins A. Differentiating between the possibility and probability of SARS-CoV-2 transmission associated with wastewater: empirical evidence is needed to substantiate risk. FEMS Microbes. 2021;2 doi: 10.1093/femsmc/xtab007. xtab007Doi. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

